License Plates : automated short RNA nomenclature
The "License Plates" nomenclature defines universal, unique, unambiguous, and reversible names for short RNA molecules. Because license plates are derived directly from the nucleotide sequence, they are independent of model organism and genome assembly and persist over time. License plates ensure that different researchers use the same name to refer to the same short RNA molecule without requiring that the researchers communicate with one another ahead of time or even know about one another's research work. License plates make the searching through and comparison of different publications easy!
There is a need for an automated and unified short RNA nomenclature
Many nomenclatures exist for different types of short RNAs, e.g., the miRBase nomenclature and the miRCarta nomenclature for naming microRNAs (miRNAs). The current naming mechanisms typically require a brokering entity (group or organization) that issues the labels, are not reversible (one has to consult an index to retrieve the molecule of interest), is not unique (one molecule has multiple names), and is not universal (molecules across assemblies and species have different names). These characteristics inhibit cross-team communication, use of existing literature and, complicate analysis pipelines.
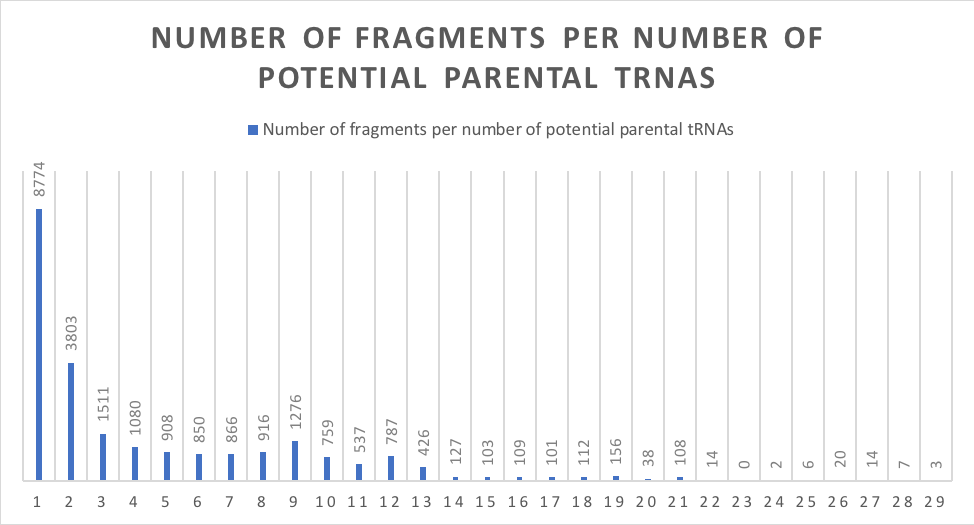
Imagine that many papers describe the same molecule. How will a researcher know this? If the name used is unique and unambiguous, all publications will use the same name; however, this is impossible if the name contains non-unique structural elements. An example of such a structural element is the tRF isodecoder. If the tRNA isodecoder is part of a tRF name, many tRFs will have multiple names because one tRF molecule can originate from many different tRNAs that sometimes have different isodecoders (image 1). In other words, multiple different labels will refer to the same exact molecule, making reading the literature more difficult.
When multiple brokering entities exist for one RNA type or a new assembly is released, or when a researcher tries to review a molecule across species, the lack of universality of nomenclature also creates issues. Often nomenclatures use arbitrary numbers (e.g., miR-1, miR-2, miR-3). Naturally, the numbers are not preserved across assemblies and species because those numbers do not have any significance. For example, miRNA names changed across miRBase versions [1]. Similarly, different brokering entities may end up using different arbitrary names for the same short RNA molecule - for example, hsa-834-875.1 (miRCarta notation) and hsa-miR-122 (miRBase notation) refer to the same parental molecule. This inconsistency makes reviews across teams, assemblies, and species difficult for the researcher.
Reversibility becomes increasingly more important the more automated analysis pipelines become. For example, if a researcher uses multiple short RNA types, they have to keep up-to-date name-to-sequence indexes (most likely large fasta files) for all the types of molecules. Each time a molecule was referenced by name, they have to look it up in the appropriate table. That process is prone to errors is time-consuming.
Finally, arguably the most complicating factor here is the dependence on a "brokering mechanism:" a dedicated group or organization curates the data, issues and communicates labels for newly discovered RNAs, and continuously updates tables that link labels and sequences by processing newly-discovered or withdrawn sequences. This approach requires the continuous availability of funds and domain experts. More to the point, the labels cannot exist until after the sequence of the corresponding RNA is published: in the meantime, the researcher has to give their newly discovered sequence an arbitrary name. It is easy to see that as new sequences appear in the literature at an increasing pace, it will prove increasingly difficult for the organizations that curate the data to keep up and generate new labels in lockstep with the literature.
License plates are a powerful, no-cost, worry-free short RNA nomenclature
We proposed the license plate nomenclature to tackle the above complications. This labeling scheme considers only the short RNA's nucleotide sequence: this results in a short RNA nomenclature that is unique, unambiguous, universal, reversible and, does not require any coordination with any brokering entity (including our team). In other words, each short RNA has its unique license plate and, each license plate corresponds to a unique short RNA sequence across all assemblies, species and, short RNA types.
License plates were described initially in the context of tRFs and deployed in MINTbase, or a database of tRFs [2,3]. Subsequently, we extended them to rRNA-derived fragments [4]. More recently, we also extended them to isomiRs [5]. It is worth noting that the recently proposed standard adopts the license plate labeling scheme for labeling, reporting, and comparing isomiRs [6].
What is a license plate for a short RNA?
The license plate nomenclature provides names to short RNAs. The new name is determined by rewriting the short RNA's nucleotide sequence in a base-32 numbering scheme (see Encoding below), unambiguous and reversible (see Decoding below). The numbering scheme uses the digits 0 through 9 and the 22 upper case letters of the English alphabet that remain after excluding A, C, G, and T [2]. The resulting license plate comprises three parts ( image 2):
- a 3-letter prefix that indicates the RNA class to which the short RNA belongs (e.g., "iso-" for the microRNA isoforms also known as isomiRs, "tRF-" for tRNA-derived fragments, and "rRF-" for rRNA-derived fragments)
- the length of the original nucleotide sequence and,
- the base-32 number.

Why use the "License Plates"?
- They obviate the need for a centralized brokering service that hands out labels. Researchers at different centers can generate the same name for the same short RNA sequence on their own and use it immediately in a manuscript. A unified and universal short RNA nomenclature ensures that different groups refer to the same short RNA using the same name in their publications. Furthermore, it ensures that searching across publications for a molecule is possible.
- The license plate of a short RNA does not change over time. I.e., the license plate is not affected by genome assembly edits or changes to the miRNA, tRNA, rRNA reference sequences.
- License plates allow uniform indexing of isomiR, tRF or, rRF, and essentially any other RNA type. Therefore, analyses across RNA types are more straightforward and faster.
- License plates facilitate frustration-free, inter-laboratory communication. Scientists from different teams can share data quickly and easily with one another without worrying about redundancies or inconsistencies.
More details: derive a License Plate for a given short RNA ("Encoding")
The naming system that we created essentially compresses the nucleotide sequence of a molecule to an alphanumeric sequence. To achieve the smaller size, instead of the 4 letters A, C, G, T, we use the English alphabet and the numbers to describe a sequence, B, D, 0, E, F, 1, H, I, 2, J, K, 3, L, M, 4, N, O, 5, P, Q, 6, R, S, 7, U, V, 8, W, X, 9, Y, Z (we do not re-use A, C, G, T for simplicity). In other words, we go from a base-4 system to a base-32 system. The fragment length is a necessary part of the name that helps with the Encoding and provides helpful information. The new label is a new sequence that resembles an automobile's license plate. Hence, the name!
Additionally, we can add a prefix to the sequence to indicate the type of the fragment. The user can select the prefix of their choice in the downloadable code or view the prefix we chose based on published data and specific types online. The prefix does not affect Encoding or Decoding and is optional.
For example, the 24 nucleotides long, GRCh37, 3'-tRF GTTCGATTCCCGGTCAGGGAACCA can originate from 13 distinct tRNAs. Those tRNAs come from three different Amino acid/Anticodons, GluCTC, PheGAA, and GluTTC, that reside in 3 different chromosomes. According to our mechanism, GTTCG=7S, ATTCC=IR, CGGTC=MM, AGGGA=12, and ACCA=E2. Therefore, the sequence's Encoding is 24-7SIRMM12E2 and the prefix GRCh37-tRF. So, the license plate of GTTCGATTCCCGGTCAGGGAACCA is tRF-24-7SIRMM12E2.
More details: retrieve the short RNA from its License Plate ("Decoding")
One can recover the initial nucleotide sequence from a given license plate using the Encoding's rules in reverse.
For example, the Decoding of the short RNA name tRF-24-7SIRMM12E2 is GTTCGATTCCCGGTCAGGGAACCA.
Let us examine how we would recover the short RNA sequence whose license plate is tRF-24-7SIRMM12E2. For this specific example, 7S= GTTCG, IR=ATTCC, MM=CGGTC, and 12=AGGGA. Note that we have recovered 20 of the 24 letters of the RNA's nucleotide sequence so far. The remaining part of the label, E2, can have multiple interpretations, as do a few other character combinations. E2 can encode ACCA and ACGGA. However, we know that there are only 4 letters left (24 nts of the initial length minus 20 nts that we have already Decoded), so E2 must be ACCA. In conclusion, tRF-24-7SIRMM12E2 corresponds to GTTCGATTCCCGGTCAGGGAACCA (image 3).
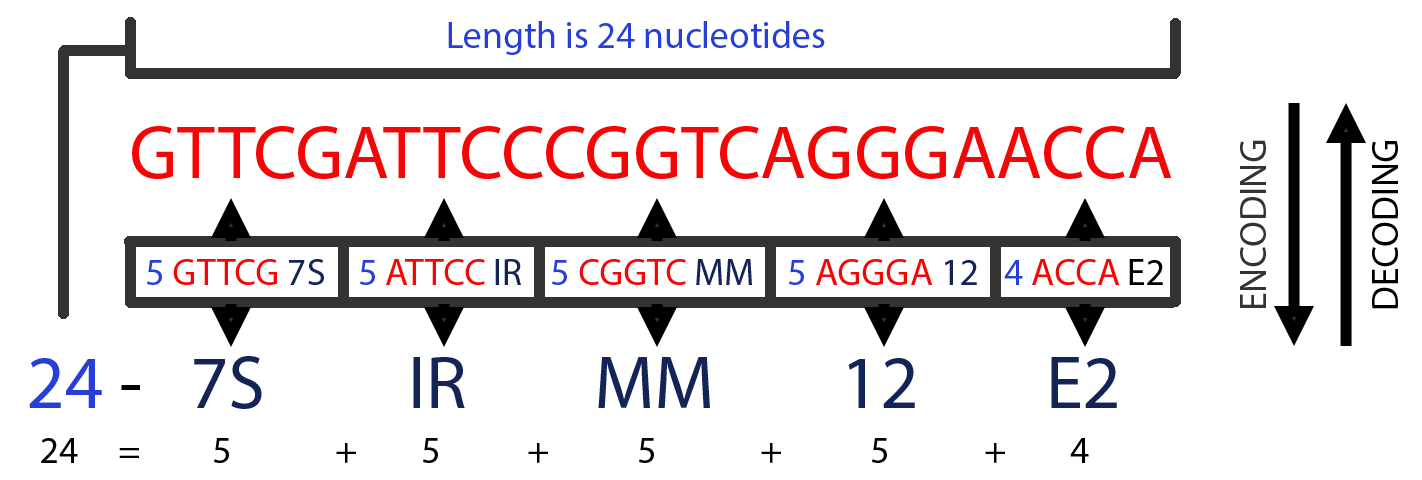
References
- Backes, C, Fehlmann, T, Kern, F, Kehl, T, Lenhof, HP, Meese, E, Keller, A. miRCarta: a central repository for collecting miRNA candidates. Nucleic Acids Res. 2018 Jan 4;46(D1):D160-D167. doi: 10.1093/nar/gkx851. PubMed PMID:29036653; PubMed Central PMCID:PMC5753177.
- Pliatsika, V, Loher, P, Telonis, AG, Rigoutsos, I. MINTbase: a framework for the interactive exploration of mitochondrial and nuclear tRNA fragments. Bioinformatics. 2016;32 (16):2481-9. doi: 10.1093/bioinformatics/btw194. PubMed PMID:27153631 PubMed Central PMC4978933.
- Pliatsika, V, Loher, P, Magee, R, Telonis, AG, Londin, E, Shigematsu, M, Kirino, Y, Rigoutsos, I. MINTbase v2.0: a comprehensive database for tRNA-derived fragments that includes nuclear and mitochondrial fragments from all The Cancer Genome Atlas projects. Nucleic Acids Res. 2018;46 (D1):D152-D159. doi: 10.1093/nar/gkx1075. PubMed PMID:29186503 PubMed Central PMC5753276.
- Cherlin, T, Magee, R, Jing, Y, Pliatsika, V, Loher, P, Rigoutsos, I. Ribosomal RNA fragmentation into short RNAs (rRFs) is modulated in a sex- and population of origin-specific manner. BMC Biol 18, 38 (2020). doi: 10.1186/s12915-020-0763-0. PubMed PMID: 32279660.
- Loher P, Karathanasis N, Londin E, Bray P, Pliatsika V, Telonis AG, Rigoutsos I. IsoMiRmap-fast, deterministic, and exhaustive mining of isomiRs from short RNA-seq datasets. Bioinformatics. 2021 Jan 20:btab016. doi: 10.1093/bioinformatics/btab016. Epub ahead of print. PubMed PMID:33471076.
- Desvignes, T, Loher, P, Eilbeck, K, Ma, J, Urgese, G, Fromm, B, Sydes, J, Aparicio-Puerta, E, Barrera, V, Espín, R, Thibord, F, Ros, XB, Londin, E, Telonis, AG, Ficarra, E, Friedländer, MR, Postlethwait, JH, Rigoutsos, I, Hackenberg, M, Vlachos, IS, Halushka, MK, Pantano, L. Unification of miRNA and isomiR research: the mirGFF3 format and the mirtop API. Bioinformatics. 2019 Aug 29. doi: 10.1093/bioinformatics/btz675. PubMed PMID:31504201.
- Magee, R, Rigoutsos, I. On the expanding roles of tRNA fragments in modulating cell behavior. Nucleic Acids Res. 2020 Sep 25;48(17):9433-9448. doi: 10.1093/nar/gkaa657. PubMed PMID:32890397.
