1. Ribosomal RNAs
Ribosomal RNAs (rRNAs) are the most abundant class of RNAs and serve as the scaffold for ribosomes, the cellular factories that ‘translate’ messenger RNAs (mRNAs) into proteins.
At any given moment, there are thousands of active RNA molecules in any one of our cells. RNAs are produced from genomic DNA and, working together with proteins, maintain cell health (“homeostasis”). Many distinct classes of RNAs, short and longer ones, have been characterized already. Each of these classes carries out its own distinct and important roles. rRNAs make up roughly 80% of all RNA found in a cell.
There are six different rRNAs: 5S, 5.8S, 12S, 16S, 18S, and 28S 1. The label of each rRNA reflects the time it takes to ‘settle’ at the bottom of a solution. The time is expressed in Svedberg (S) units: the larger the S-value of an rRNA the larger its mass and the longer it takes to settle. rRNAs with different S-values differ not only in mass but also in sequence composition and shape 2. Figure 1 shows the secondary structure of the 5.8S rRNA 3.
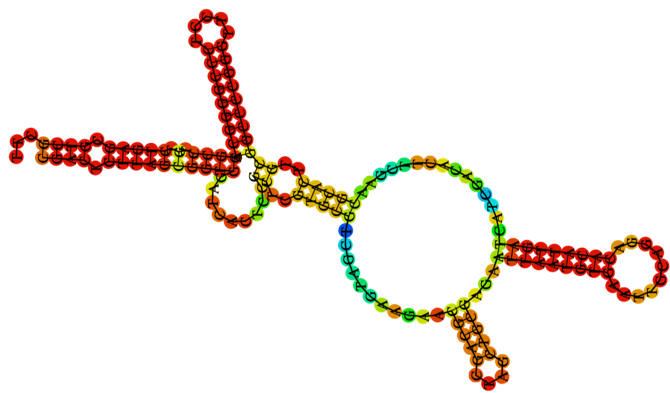 Figure 1: The secondary structure of the 5.8S rRNA 3
Figure 1: The secondary structure of the 5.8S rRNA 3
Four of the six rRNAs (5S, 5.8S, 18S, and 28S) are located in the nuclear genome. The remaining two rRNAs (12S and 16S) are located on the circular mitochondrial (MT) genome where they are flanked by MT tRNAs phenylalanine, valine, and leucine 1,4.
Three of the four nuclearly-encoded rRNAs (5.8S, 18S, and 28S) exist as a gene cluster or “cassette” known as “45S.” The encodings for the 45S clusters are found on the p-arms of the acrocentric chromosomes 13, 14, 15, 21, and 22 5,6. The encodings for 5S are found primarily as tandem repeats on chromosome 1 7,8. The 45S and 5S genes are estimated to have a combined total of many hundreds of copies in the nuclear genome. The high number of copies is meant to facilitate rapid and frequent transcription of the nuclear rRNAs that are needed for protein synthesis 7,9,10.
Once transcribed, the 45S is processed into the 5.8S, 18S, and 28S rRNAs and begins complexing with ribosomal proteins as they are exported from the nucleus into the cytoplasm 1,11. The 5S, 5.8S, and 28S combine to form the scaffold for the large ribosomal subunit (LSU) 1,11. The 18S is the sole scaffolding rRNA for the small ribosomal subunit (SSU) 11. In the cytoplasm the ribosome complex completes its formation 5.
The two MT rRNAs are produced differently. The entire MT genome is transcribed in a polycystronic fashion into one long RNA that is then processed into individual RNAs 4,12. After being processed, the 16S becomes the scaffold for the MT LSU component of the MT ribosome while the 12S becomes the scaffold for the MT SSU component of the MT ribosome 12. While there exists only one copy of 12S and 16S rRNA gene in the MT genome, there are many mitochondria (100s to 1000s) in a cell, and thus, many copies of the MT genome from which 12S and 16S rRNAs are made 13,14. The varying numbers of mitochondria and MT genome copies in a cell mean that the number of MT rRNAs will vary from cell to cell and person to person 13,14.
2. Ribosomal RNA fragments (rRFs)
rRNA-derived fragments (rRFs) are short non-coding RNAs (ncRNAs) that are derived from both nuclear and mitochondrial rRNAs22. These fragments are often abundant and shown to be modulated in both a sex and population dependent manner.
Next generation sequencing (NGS) is a powerful tool for RNA discovery and has made it possible for scientists to catalogue the RNAs in our cells in an unbiased way. Many important long and short RNAs have been discovered and characterized in the context of health and disease. It had been known for several years that small noncoding RNAs (sncRNAs) like microRNAs (miRNAs), miRNA isoforms (isomiRs), and tRNA-derived fragments (tRFs) have functional roles in the regulation of gene expression. The Rigoutsos lab showed that both miRNAs and tRNAs produce in a regimented manner “clouds” of co-existing isomiRs 15 and tRFs 16. IsomiRs and tRFs from the same parental template typically share the same core sequence and differ in their endpoints. The Rigoutsos lab also showed that the identities and abundances of isomiRs and tRFs depend on personal attributes like sex, population origin, and race/ethnicity as well as on tissue type, tissue state, and disease type 17–21.
A recent publication from the Rigoutsos lab characterized the rRFs that are produced from the six rRNAs (Figure 2)22. They analyzed the sncRNA profiles of lymphoblastoid cell lines derived from 434 participants to the 1000 Genomes Project. The participants comprised healthy men and women from five geographical populations. It was our goal to understand both the universal and population-specific profiles of rRFs. We showed that rRNAs produce “clouds” of rRFs in a regimented manner. We also showed that there are thousands of rRFs produced from specific locations across all six rRNAs. These rRFs have unique length and abundance profiles. Our analyses showed that many of these rRFs are ubiquitous and present in all analyzed samples. However, we also found a significant number of rRFs that are dependent on the donors’ personal attributes including sex and population origin, as well as on tissue type and disease. These properties mirror the lab’s earlier findings on isomiRs and tRFs.
 Figure 2: The four nuclear and two mitochondrial rRNAs produce many fragments (rRFs) in a regimented manner
Figure 2: The four nuclear and two mitochondrial rRNAs produce many fragments (rRFs) in a regimented manner
rRFs are the newest addition to the sncRNA repertoire. They represent the third class of sncRNAs whose production depends on personal attributes and cellular context. The manner in which rRFs are produced is not known. Also not known are the rRFs’ mechanism of action and functional roles. These questions are currently an area of active investigation by the researchers in Jefferson’s Computational Medicine Center.
References
- Tafforeau, L, Zorbas, C, Langhendries, JL, Mullineux, ST, Stamatopoulou, V, Mullier, R, et al. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of pre-rRNA processing factors. Molecular Cell. 2013;51:539-51. doi: 10.1016/j.molcel.2013.08.011.
- Lehninger, AL, Cox, DL, Nelson, MM. Principles Of Biochemistry. New York, NY: Worth Publishers, 1993. 2nd edition.
- Hofacker IL. Vienna RNA secondary structure server. Nucleic acids research. 2003;31:3429-31. doi: 10.1093/nar/gkg599.
- De Silva, D, Tu, Y-T, Amunts, A, Fontanesi, F, Barrientos, A. Mitochondrial ribosome assembly in health and disease. Cell Cycle. 2015;14:2226-50. doi: 10.1080/15384101.2015.1053672.
- Boisvert, F-M, van Koningsbruggen, S, Navascués, J, Lamond, AI. The multifunctional nucleolus. Nature Reviews Molecular Cell Biology. 2007;8:574-85. doi: 10.1038/nrm2184.
- Henderson, AS, Warburton, D, Atwood, KC. Location of ribosomal DNA in the human chromosome complement. Proc Natl Acad Sci U S A. 1972;69:3394-8. doi: 10.1073/pnas.69.11.3394.
- Yu, S, Lemos, B. A Portrait of Ribosomal DNA Contacts with Hi-C Reveals 5S and 45S rDNA Anchoring Points in the Folded Human Genome. Genome Biol Evol. 2016;8:3545-58. doi: 10.1093/gbe/evw257.
- Akamatsu, Y, Kobayashi, T. The Human RNA Polymerase I Transcription Terminator Complex Acts as a Replication Fork Barrier That Coordinates the Progress of Replication with rRNA Transcription Activity. Molecular and Cellular Biology. 2015;35:1871-81. doi: 10.1128/MCB.01521-14.
- Karahan, G, Sayar, N, Gozum, G, Bozkurt, B, Konu, O, Yulug, IG. Relative expression of rRNA transcripts and 45S rDNA promoter methylation status are dysregulated in tumors in comparison with matched-normal tissues in breast cancer. Oncology Reports. 2015;33:3131-45. doi: 10.3892/or.2015.3940.
- Sorensen, PD, Frederiksen, S. Characterization of human 5S rRNA genes. Nucleic Acids Res. 1991;19:4147-51. doi: 10.1093/nar/19.15.4147.
- Ciganda, M, Williams, N. Eukaryotic 5S rRNA biogenesis. Wiley Interdiscip Rev RNA. 2011;2:523-33. doi: 10.1002/wrna.74.
- Christianson, TW, Clayton, Da. A tridecamer DNA sequence supports human mitochondrial RNA 3′-end formation in vitro. Molecular and cellular biology. 1988;8:4502-9. doi: 10.1128/MCB.8.10.4502.Updated.
- Bogenhagen, D, Clayton, DA. The number of mitochondrial deoxyribonucleic acid genomes in mouse L and human HeLa cells. Quantitative isolation of mitochondrial deoxyribonucleic acid. J Biol Chem. 1974;249:7991-5.
- Moraes, C. What regulates mitochondrial DNA copy number in animal cells? Trends in Genetics. 2001;17:199-205. doi: 10.1016/S0168-9525(01)02238-7.
- Loher P, Londin ER, Rigoutsos I. IsomiR expression profiles in human lymphoblastoid cell lines exhibit population and gender dependencies. Oncotarget. 2014;5:8790-802. doi: 10.18632/oncotarget.2405.
- Telonis, AG, Loher, P, Honda, S, Jing, Y, Palazzo, J, Kirino, Y, et al. Dissecting tRNA-derived fragment complexities using personalized transcriptomes reveals novel fragment classes and unexpected dependencies. Oncotarget. 2015;6:24797-822. doi: 10.18632/oncotarget.4695.
- Telonis, AG, Loher, P, Jing, Y, Londin, E, Rigoutsos, I. Beyond the one-locus-one-miRNA paradigm: microRNA isoforms enable deeper insights into breast cancer heterogeneity. Nucleic Acids Res. 2015;43:9158-75. doi: 10.1093/nar/gkv922.
- Telonis, AG, Magee, R, Loher, P, Chervoneva, I, Londin, E, Rigoutsos, I. Knowledge about the presence or absence of miRNA isoforms (isomiRs) can successfully discriminate amongst 32 TCGA cancer types. Nucleic Acids Res. 2017;45:2973-85. doi: 10.1093/nar/gkx082.
- Pliatsika, V, Loher, P, Magee, R, Telonis, AG, Londin, E, Shigematsu, M, et al. MINTbase v2.0: a comprehensive database for tRNA-derived fragments that includes nuclear and mitochondrial fragments from all The Cancer Genome Atlas projects. Nucleic Acids Res. 2018;46:D152-D9. doi: 10.1093/nar/gkx1075.
- Magee, RG, Telonis, AG, Loher, P, Londin, E, Rigoutsos, I. Profiles of miRNA Isoforms and tRNA Fragments in Prostate Cancer. Sci Rep. 2018;8:5314. doi: 10.1038/s41598-018-22488-2.
- Telonis, AG, Loher, P, Magee, R, Pliatsika, V, Londin, E, Kirino, Y, et al. tRNA Fragments Show Intertwining with mRNAs of Specific Repeat Content and Have Links to Disparities. Cancer Res. 2019.
- Cherlin, T, Magee, R, Jing, Y, Pliatsika, V, Loher, P, Rigoutsos, I. Ribosomal RNA fragmentation into short RNAs (rRFs) is modulated in a sex- and population of origin-specific manner. BMC Biol 18, 38 (2020). doi: 10.1186/s12915-020-0763-0. PubMed PMID:32279660.
