Commercial quantitative polymerase chain reaction (qPCR) methods are well employed for the study and characterization of short non-coding RNAs such as microRNAs (miRNAs). Though these methods rely discriminate individual miRNAs, less is known about their capacity to discriminate isomiRs with closely related sequences.
In our recent work1, we investigate the ability of two commonly employed methods – ThermoFisher Taqman qPCR and Exiqon miRCuRY LNA qPCR – to sensitively and specifically detect four isomiRs of hsa-miR-21-5p in both synthetic contexts and real cell contexts. Using synthetic RNA oligonucleotides and transfection of miRNA mimics, we questioned whether either of these methods could adequately resolve changes in isomiR concentrations in both the presence and absence of closely related isomiRs.
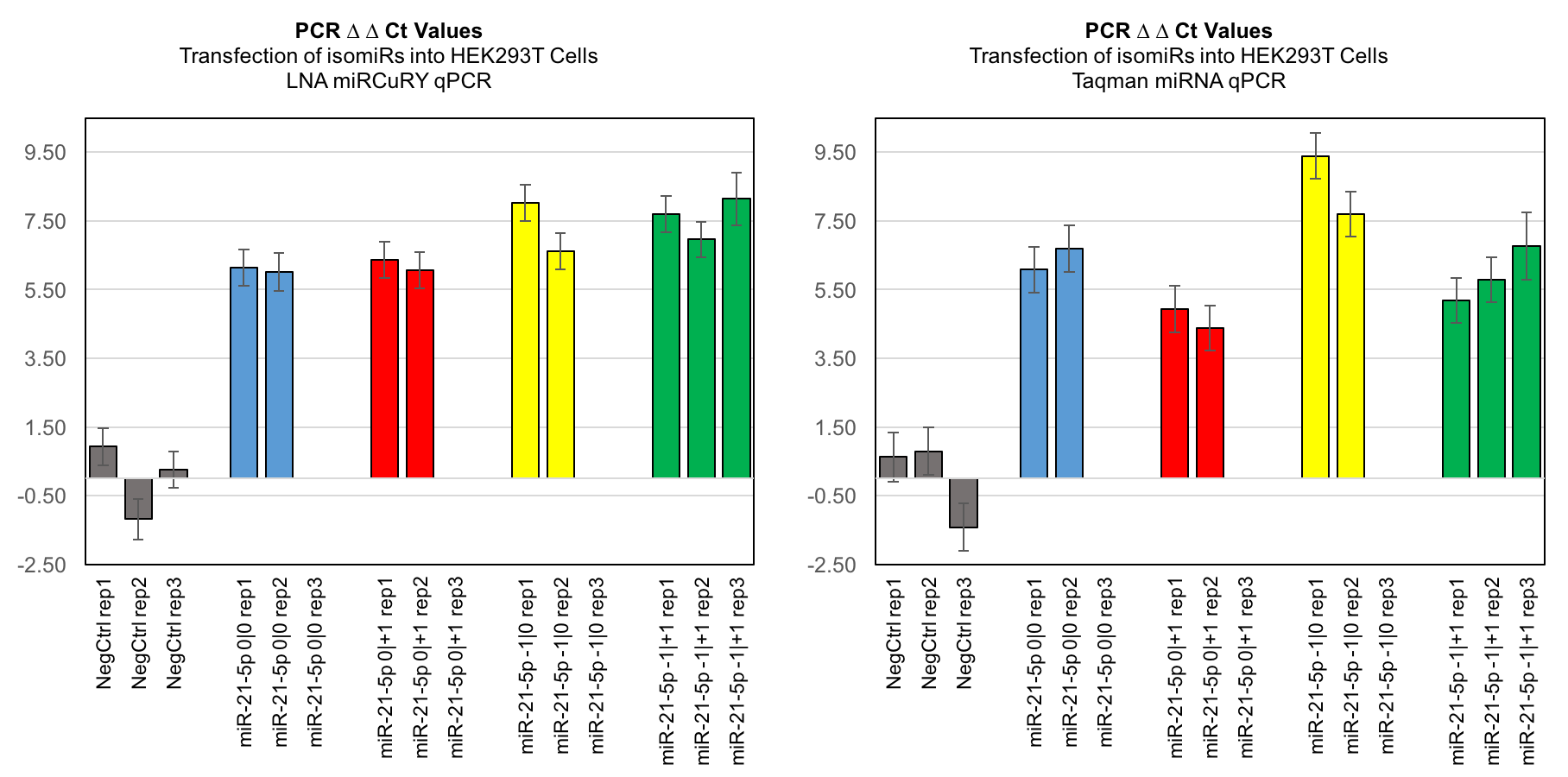 Comparing the transfection of isomiRs into HEK293T Cells in LNA miRCuRY qPCR and Taqman miRNA qPCR
Comparing the transfection of isomiRs into HEK293T Cells in LNA miRCuRY qPCR and Taqman miRNA qPCR
Our results indicate that these methods exhibit significant background “cross talk”, in that closely related isomiRs do contribute background signals in qPCR. This should be taken into consideration in the design of studies on isomiRs and other short non-coding RNAs that show closely related sequences.
References
- Magee, R, Telonis, AG, Cherlin, T, Rigoutsos, I, Londin, E. Assessment of isomiR Discrimination Using Commercial qPCR Methods. Non-Coding RNA 2017, 3(2), 18; doi:10.3390/ncrna3020018
