The Mitochondrial and Nuclear rRNA fragment database (MINRbase), is a database facilitating the study of ribosomal RNA-derived fragments (rRFs). The database provides 130 238 expressed rRFs arising from human nuclear rRNAs (18S, 5.8S, 28S, 5S), two mitochondrial rRNAs (12S, 16S) or four spacers of 45S pre-rRNA, all of which were compiled from 11 632 datasets.
MINRbase allows the user to perform complex queries using various filters and criteria such as parental rRNA identity, nucleotide sequence, rRF minimum abundance, and metadata keywords (e.g. tissue type, disease), all through its easy-to-use user interface. A ‘summary’ page is provided for each rRF, granting the user with a detailed breakdown of the expression by tissue type, disease, sex, ancestry and other variables; it also allows the user to generate publication-ready plots with a click of a button.
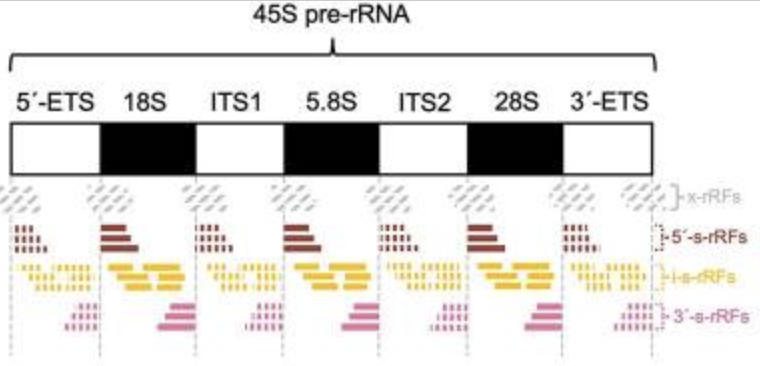
MINRbase has provided support for novel observations, such as the internal spaces of 45S are prolific producers of abundant rRFs and many abundant rRFs straddle the known boundaries of rRNAs. MINRbase can be accessed at https://cm.jefferson.edu/MINRbase/
References
- Pliatsika, V, Cherlin, T, Loher, P, Vlantis, P, Nagarkar, P, Nersisyan, S, Rigoutsos, I. MINRbase: a comprehensive database of nuclear- and mitochondrial-ribosomal-RNA-derived fragments (rRFs). Nucleic Acids Res. 2023; :. doi: 10.1093/nar/gkad833. PubMed PMID:37843123.
