Genes and genomes are structured in very complex ways. The vast majority of protein-coding genes contain introns: some, like UMPS, contain a couple of introns while others, like MAP3K4, can contain tens of introns. In addition, the content of these genes in repetitive elements greatly differs: about 25% of UMPS’ body is Alu elements, while only 6% of MAP3K4 overlaps Alu elements. Our newly published research shed lights on the dynamics of gene expression during early development and uncovers common characteristics in gene expression changes.
We find common characteristics of the genes who increase in expression as development progresses from the zygote to the blastocyst. These genes are short and dense in repetitive elements, most prominently in Alu elements. On the other hand, the genes that decrease are long and sparse in repetitive elements. In fact, we were able to find unique pyknons (motifs in the DNA sequence) of each class of genes. This was a noteworthy distinction as it now helps us classify genes not only based on their function but also based on their structure. Thus, from a developmental perspective, cells seem to utilize pyknons and repetitive elements as ‘bookmarks’ on the genome that can help guide the access to certain genes when they want to quickly proliferate.
Mining additional data, we examined how pluripotency and differentiated tissues fit into the observed dichotomies. Our findings revealed that pluripotency includes short and Alu-rich genes while the tissue-specific genes are long and sparse in repetitive elements.
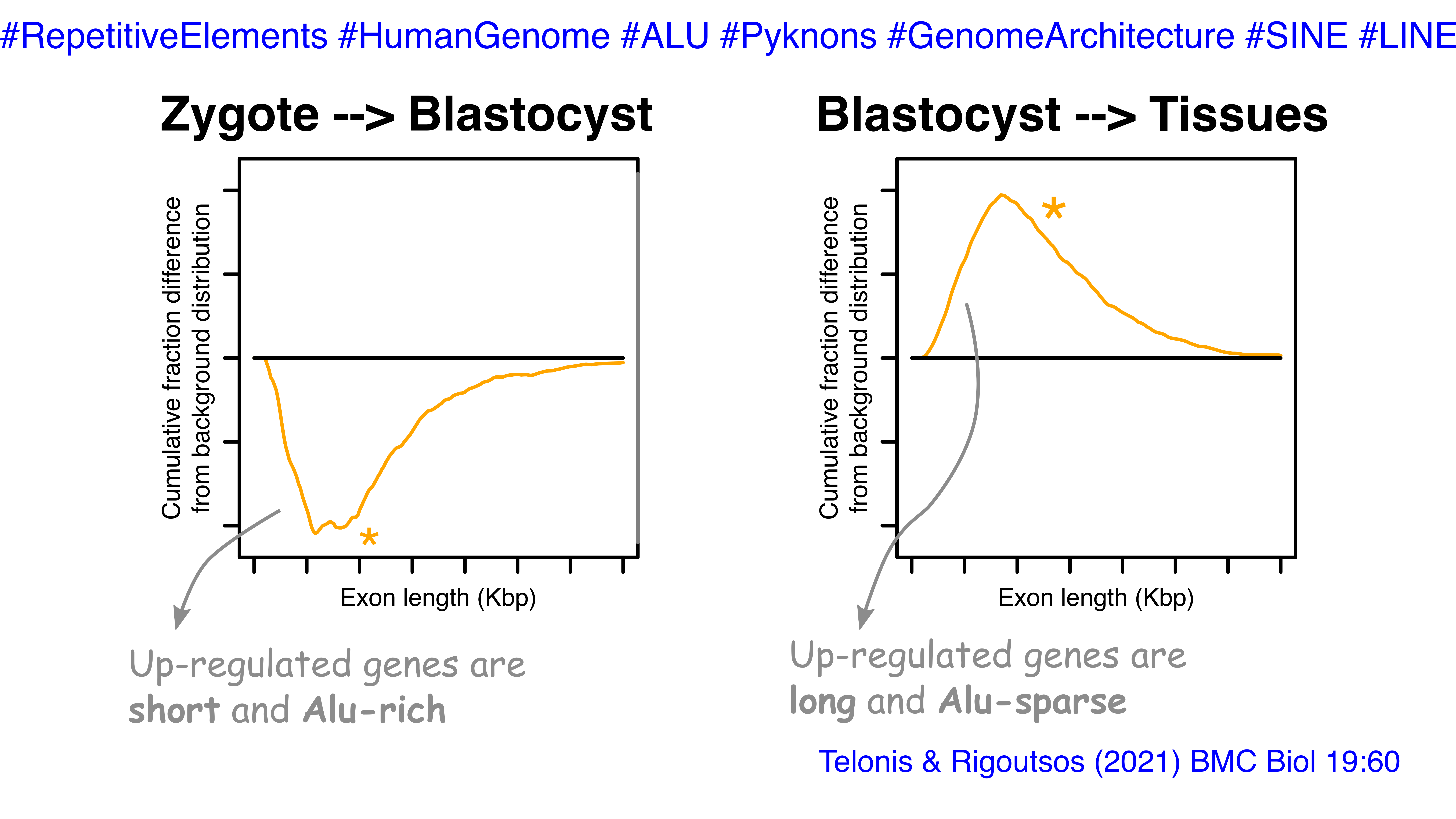 Genes with highest expression at the blastocyst stage are short and dense in Alu elements. In our most recent manuscript, we show that this observation correlated with the pluripotency/proliferation status of the cells. We also find that tissue specificity in RNA expression is associated with long genes sparse in repetitive elements.
Genes with highest expression at the blastocyst stage are short and dense in Alu elements. In our most recent manuscript, we show that this observation correlated with the pluripotency/proliferation status of the cells. We also find that tissue specificity in RNA expression is associated with long genes sparse in repetitive elements.
Our findings describe developmental processes from a novel perspective beyond transcription factors: the common denominator in expression changes is the underlying architecture and content in repetitive elements.
References
- Telonis AG, Rigoutsos I. The transcriptional trajectories of pluripotency and differentiation comprise genes with antithetical architecture and repetitive-element content. BMC Biol. 2021 Mar 25;19(1):60. doi: 10.1186/s12915-020-00928-8. PubMed PMID:33765992.
