Over the last few years, extracellular RNAs (exRNAs) in biofluids has gained increased attention for their potential diagnostic, prognosis, and monitoring therapeutic potential. Despite the value of exRNAs, the field has been hampered by inter- and intra-study reproducibility, possible due to technical and biological variations, making it difficult to reach a clinical utility of the different biomarkers. This is particularly important for molecules that rely on differences in abundances, such as RNAs, rather than the detection of disease-associated mutations, which may be more tolerant to technical variation. One source of variability, is the technical variations introduced dues to the methods of exRNA isolation and RNA profiling. As exRNAs can be associated with subclasses including extracellular vesicles (EVs), ribonucleoprotein complexes (RNPs), and lipoprotein complexes (LPPs), all of which carry different RNA and protein cargo and present in different concentrations based upon source of cell type, environmental, epigenetic, and other factors. Therefore, approaches to isolate exRNAs differentially isolate the various carrier subclasses and therefore measure different molecules, leading to a source of variability.
In this collaborative study addressed this issue, by performing a systematic investigation of 10 exRNA isolation methods across five biofluids and cell lines, with the goal of addressing the technical variation introduced by these methods and improve inter-study reproducibility. Specifically, this comprehensive study focused on the profiles of the short non-coding exRNAs following the various isolation methods. This study produced key findings that can be used to guide future exRNA biomarker identification studies. Importantly, the performance of exRNA isolation used varied across biological fluids and RNA species types resulting in differences in reproducibility rates.
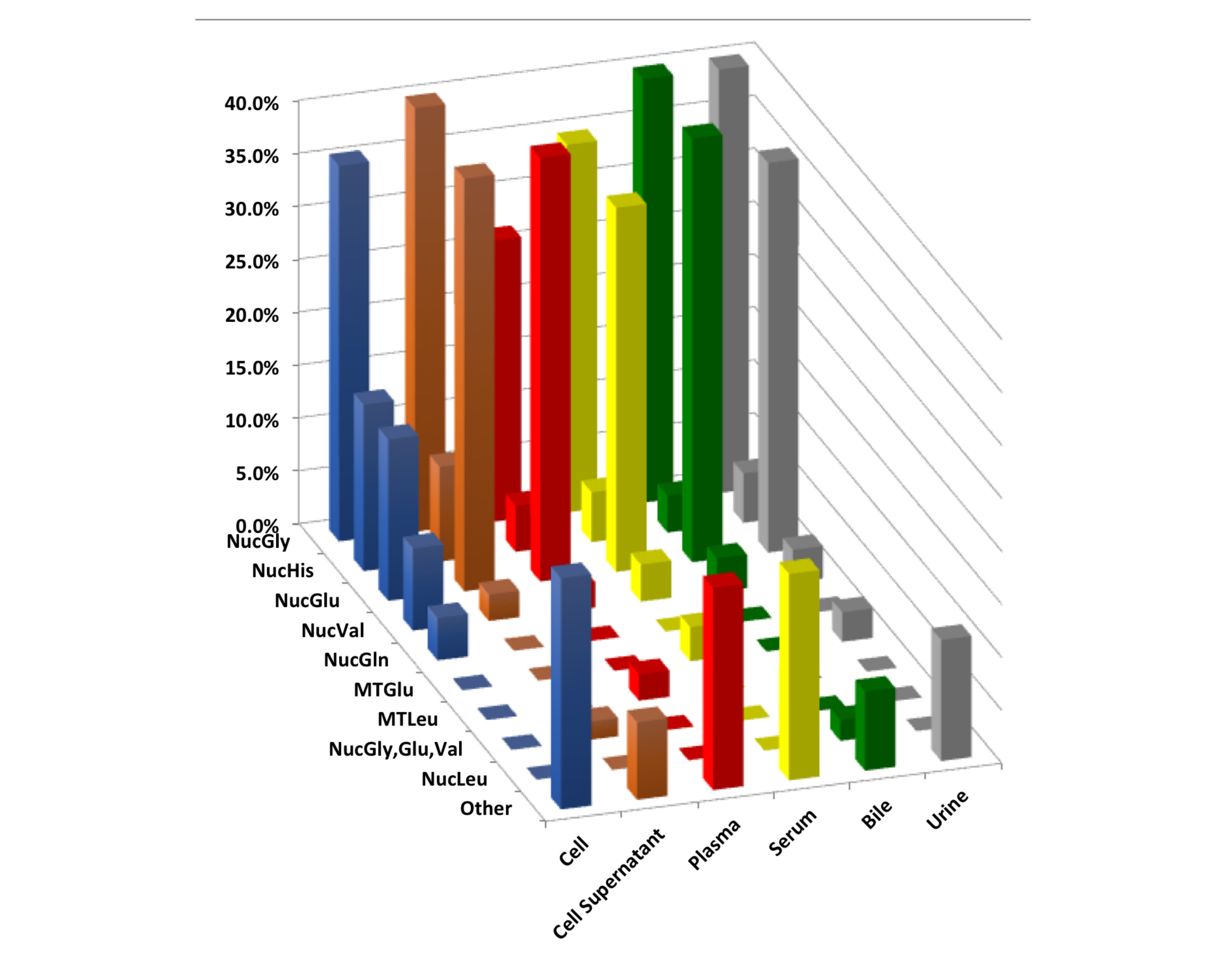
MiRNA profiles had marked differences in profiles due to a varied of variables including sequence read depth, exRNA isolation method, and biofluid. Specifically, the various exRNA isolation methods results in substantial differences in miRNA profiles for bile, plasma, and serum biofluids, but not for urine of cell culture condition medium (CCCM). Similarly, tRNA derived fragments (tRFs) revealed marked differences in profiles across the different biofluid type and cell lines examined. Yet, the distribution of tRF sizes and distribution of amino acid preferences were similar across exRNA isolation methods. The impact of exRNA isolation was limited to an increase in the fraction of 3’-tRFs within the plasma samples. Overall, the impact of exRNA isolation method had a weaker effect on tRNA profiles than the miRNA profiles. These observations suggest a utility for tRFs as exRNA biomarkers that show less technical variability amongst isolation approaches than miRNAs.
This comprehensive study sets the groundwork for future exRNA biomarker discovery efforts. Through a rigorous comparisons of different exRNA biofluids and isolation techniques, major sources of the technical and biological variability that has hampered the field have been identified.
References
- Srinivasan, S, Yeri, A, Cheah, PS, Chung, A, Danielson, K, De Hoff, P, Filant, J, Laurent, CD, Laurent, LD, Magee, R, Moeller, C, Murthy, VL, Nejad, P, Paul, A, Rigoutsos, I, Rodosthenous, R, Shah, RV, Simonson, B, To, C, Wong, D, Yan, IK, Zhang, X, Balaj, L, Breakefield, XO, Daaboul, G, Gandhi, R, Lapidus, J, Londin, E, Patel, T, Raffai, RL, Sood, AK, Alexander, RP, Das, S, Laurent, LC. Small RNA Sequencing across Diverse Biofluids Identifies Optimal Methods for exRNA Isolation. Cell. 2019;177 (2):446-462.e16. doi: 10.1016/j.cell.2019.03.024. PubMed PMID:30951671 PubMed Central PMC6557167.