For a quickstart guide, go directly to the “execution examples and what to use when” section | Try our new Teiresias web application
If you use any of the below please cite: Rigoutsos, I, Floratos, A (1998) Combinatorial pattern discovery in biological sequences: The TEIRESIAS algorithm. Bioinformatics 14: 55-67
General
What is Teiresias
The Teiresias algorithm is a combinatorial algorithm for the discovery of rigid patterns (motifs) in biological sequences. It is named after the Greek prophet Teiresias and was created in 1997 by Isidore Rigoutsos and Aris Floratos. Teiresias should be used for the discovery of exact or more lenient similarities within a set of character sequences. In other words, Teiresias finds all patterns that exist in the input using each input character as a unit. It was initially developed for the discovery of sequences of amino acids of sequenced data but can be used in any text mining setting. You can download the Teiresias source files for local use here and you can access our Teiresias web tool here.
Glossary
- Pattern: The Teiresias algorithm uses regular expressions to define patterns. The patterns created by the algorithm are <L,W> patterns that have at least k instances in the input, where L ≤ W and L, W, k positive integers. A pattern is called an <L,W> pattern if and only if any L consecutive literals or bracketed literals span at most W positions (i.e. there can be no more than W-L wild cards).
- Characters – Literal/Wildcard/Bracket: The algorithm returns patterns that contain a) literals, that is characters, e.g. ‘Y’, b) wild cards, that is a dot ‘.’ that represents any character of the input and c) brackets, that is literals in a bracket, e.g. [YW] that can be used interchangeably in the pattern’s appearances
- Label line: A line of the input file that starts with ‘>’. Those lines can be used to include information about each sequence and are ignored by the algorithm. Any line that shouldn’t be considered by the algorithm should start with ‘>’
- Equivalent Literals: The literals that are on the same line in the equivalence file, those literals will be considered interchangeable and will reside in the same bracket if needed
- Offset List: A list of pairs of integer numbers. The first number is the sequence number, the second number is the position in the sequence. The list accompanies each pattern and contains the sequences and positions that the pattern occupies in the input. Please remember that the numbering of the sequences and of the positions starts at 0 and not at 1
- Maximal pattern: Given a set of sequences S, a pattern P that appears k times in S is called maximal if and only if there exists no pattern P’ which is more specific than P and also appears exactly k times in S
Algorithm Guarantees
The algorithm
- Is able to produce all patterns that have a user-defined number of copies in the given input
- Manages to be very efficient by avoiding the enumeration of the entire space
- The algorithm reports motifs that are maximal in both length and composition
Source Code
Parameters
- -l: REQUIRED. This is the minimum number of literals and/or brackets of the output patterns. Every output pattern will have length at least -l and will contain at least -l literals and/or brackets. It should always be followed by a number, e.g. -l4
- -w: REQUIRED. This parameter controls the minimum literal and/or bracket density of the output patterns. In the worst case an output pattern will have length -w and -l literals and/or brackets. The rest w-l will be wildcards. It should always be at least equal to -l and followed by a number, e.g. -w6
- -k: REQUIRED. This is the minimum support that any pattern can have. Every reported pattern will have at least -k appearances in the file. It should always be larger than or qual to 2 and followed by a number, e.g. -k5
- -q: This is the maximum support that any pattern can have. Every reported pattern will have at most -q appearances in the file. By omitting it, it gets automatically set to “infinity”. It should always be followed by a number, e.g. -q85
- -v: This parameter controls how patterns occurrences are counted. If omitted then all occurrences regardless of the sequence they appear in are counted and taken into account for -k. If it is included in the input parameters then each pattern is counted once per sequence. It should be set depending on what the user is looking for. If the number of sequences containing a pattern is sought then it should be set (given as a parameter) if the user is asking how many times a pattern is present on the input file it should be omitted.
- -i: REQUIRED. The name of the input file. Check below for the correct input file format. It should always be followed by a file name, e.g. -iInput.txt
- -o: The name of the output file. If omitted the name will be “output.txt”. Check below for the correct output file format. It should always be followed by a file name or omitted, e.g. -oOUT.txt
- -b: The name of the equivalence file. If omitted no equivalences are considered. If added then all characters that are on the same line are considered equivalent and can be interchanged in patterns. Check below for the correct equivalence file format. It should always be followed by a file name, e.g. -bHomologies.txt
- -n: The number of brackets a pattern can have. If it’s not set then there is no bracket limit. Should always be followed by a number, e.g. -n10
- -p: Prints the offset lists
- -s: When this parameter is set, only patterns that satisfy the minimum requirements set by the user (elementary patterns) are returned
- -c: This parameter controls the convolution length. In this version it is set to (-l)-1 always. It can still be used but will have no effect on the output of the algorithm
- -u,-m,-d,-r: Those parameters were used in previous version and are not included in the current one for the moment. They can still be used but will have no effect on the output of the algorithm
File Formats
Please read carefully the following instructions. The correctness of your results depend on the structure of the input and equivalence files as well as on the usage of the parameters.
Input File Format
The input file must contain all data you want processed. It is assumed that every input file contains sequences and sequence headers. Starting from version 0.9.1 the headers are not required. A header is a line that starts with “>”. Please note that any line that starts with “>” will not be considered by the algorithm and every other line will.
Every line that doesn’t start with “>” is considered a separate sequence that spans from the last character of the previous header or the beginning of the file until a new header is found or the end of the file. Unlike previous versions, the input file can contain several new line characters within a sequence. Empty lines are skipped but spaces shouldn’t appear in the sequence lines. Please, keep in mind that the format of the input file is not only important for the proper execution of the program and the proper results but also the header positioning affects the -v parameter.
The input must be in ASCII format (.txt files with rich text option turned off). A sample input file with the name “SampleInputTJU.txt”, included in the downloadable files, is the following.
View sample input SampleInputTJU.txt file>sp|P38398|BRCA1_HUMAN Breast cancer type 1 susceptibility protein OS=Homo sapiens GN=BRCA1 PE=1 SV=2
MDLSALRVEEVQNVINAMQKILECPICLELIKEPVSTKCDHIFCKFCMLKLLNQKKGPSQ
CPLCKNDITKRSLQESTRFSQLVEELLKIICAFQLDTGLEYANSYNFAKKENNSPEHLKD
EVSIIQSMGYRNRAKRLLQSEPENPSLQETSLSVQLSNLGTVRTLRTKQRIQPQKTSVYI
ELGSDSSEDTVNKATYCSVGDQELLQITPQGTRDEISLDSAKKAACEFSETDVTNTEHHQ
PSNNDLNTTEKRAAERHPEKYQGSSVSNLHVEPCGTNTHASSLQHENSSLLLTKDRMNVE
KAEFCNKSKQPGLARSQHNRWAGSKETCNDRRTPSTEKKVDLNADPLCERKEWNKQKLPC
SENPRDTEDVPWITLNSSIQKVNEWFSRSDELLGSDDSHDGESESNAKVADVLDVLNEVD
EYSGSSEKIDLLASDPHEALICKSERVHSKSVESNIEDKIFGKTYRKKASLPNLSHVTEN
LIIGAFVTEPQIIQERPLTNKLKRKRRPTSGLHPEDFIKKADLAVQKTPEMINQGTNQTE
QNGQVMNITNSGHENKTKGDSIQNEKNPNPIESLEKESAFKTKAEPISSSISNMELELNI
HNSKAPKKNRLRRKSSTRHIHALELVVSRNLSPPNCTELQIDSCSSSEEIKKKKYNQMPV
RHSRNLQLMEGKEPATGAKKSNKPNEQTSKRHDSDTFPELKLTNAPGSFTKCSNTSELKE
FVNPSLPREEKEEKLETVKVSNNAEDPKDLMLSGERVLQTERSVESSSISLVPGTDYGTQ
ESISLLEVSTLGKAKTEPNKCVSQCAAFENPKGLIHGCSKDNRNDTEGFKYPLGHEVNHS
RETSIEMEESELDAQYLQNTFKVSKRQSFAPFSNPGNAEEECATFSAHSGSLKKQSPKVT
FECEQKEENQGKNESNIKPVQTVNITAGFPVVGQKDKPVDNAKCSIKGGSRFCLSSQFRG
NETGLITPNKHGLLQNPYRIPPLFPIKSFVKTKCKKNLLEENFEEHSMSPEREMGNENIP
STVSTISRNNIRENVFKEASSSNINEVGSSTNEVGSSINEIGSSDENIQAELGRNRGPKL
NAMLRLGVLQPEVYKQSLPGSNCKHPEIKKQEYEEVVQTVNTDFSPYLISDNLEQPMGSS
HASQVCSETPDDLLDDGEIKEDTSFAENDIKESSAVFSKSVQKGELSRSPSPFTHTHLAQ
GYRRGAKKLESSEENLSSEDEELPCFQHLLFGKVNNIPSQSTRHSTVATECLSKNTEENL
LSLKNSLNDCSNQVILAKASQEHHLSEETKCSASLFSSQCSELEDLTANTNTQDPFLIGS
SKQMRHQSESQGVGLSDKELVSDDEERGTGLEENNQEEQSMDSNLGEAASGCESETSVSE
DCSGLSSQSDILTTQQRDTMQHNLIKLQQEMAELEAVLEQHGSQPSNSYPSIISDSSALE
DLRNPEQSTSEKAVLTSQKSSEYPISQNPEGLSADKFEVSADSSTSKNKEPGVERSSPSK
CPSLDDRWYMHSCSGSLQNRNYPSQEELIKVVDVEEQQLEESGPHDLTETSYLPRQDLEG
TPYLESGISLFSDDPESDPSEDRAPESARVGNIPSSTSALKVPQLKVAESAQSPAAAHTT
DTAGYNAMEESVSREKPELTASTERVNKRMSMVVSGLTPEEFMLVYKFARKHHITLTNLI
TEETTHVVMKTDAEFVCERTLKYFLGIAGGKWVVSYFWVTQSIKERKMLNEHDFEVRGDV
VNGRNHQGPKRARESQDRKIFRGLEICCYGPFTNMPTDQLEWMVQLCGASVVKELSSFTL
GTGVHPIVVVQPDAWTEDNGFHAIGQMCEAPVVTREWVLDSVALYQCQELDTYLIPQIPH
SHY
>sp|P51587|BRCA2_HUMAN Breast cancer type 2 susceptibility protein OS=Homo sapiens GN=BRCA2 PE=1 SV=2
MPIGSKERPTFFEIFKTRCNKADLGPISLNWFEELSSEAPPYNSEPAEESEHKNNNYEPN
LFKTPQRKPSYNQLASTPIIFKEQGLTLPLYQSPVKELDKFKLDLGRNVPNSRHKSLRTV
KTKMDQADDVSCPLLNSCLSESPVVLQCTHVTPQRDKSVVCGSLFHTPKFVKGRQTPKHI
SESLGAEVDPDMSWSSSLATPPTLSSTVLIVRNEEASETVFPHDTTANVKSYFSNHDESL
KKNDRFIASVTDSENTNQREAASHGFGKTSGNSFKVNSCKDHIGKSMPNVLEDEVYETVV
DTSEEDSFSLCFSKCRTKNLQKVRTSKTRKKIFHEANADECEKSKNQVKEKYSFVSEVEP
NDTDPLDSNVAHQKPFESGSDKISKEVVPSLACEWSQLTLSGLNGAQMEKIPLLHISSCD
QNISEKDLLDTENKRKKDFLTSENSLPRISSLPKSEKPLNEETVVNKRDEEQHLESHTDC
ILAVKQAISGTSPVASSFQGIKKSIFRIRESPKETFNASFSGHMTDPNFKKETEASESGL
EIHTVCSQKEDSLCPNLIDNGSWPATTTQNSVALKNAGLISTLKKKTNKFIYAIHDETSY
KGKKIPKDQKSELINCSAQFEANAFEAPLTFANADSGLLHSSVKRSCSQNDSEEPTLSLT
SSFGTILRKCSRNETCSNNTVISQDLDYKEAKCNKEKLQLFITPEADSLSCLQEGQCEND
PKSKKVSDIKEEVLAAACHPVQHSKVEYSDTDFQSQKSLLYDHENASTLILTPTSKDVLS
NLVMISRGKESYKMSDKLKGNNYESDVELTKNIPMEKNQDVCALNENYKNVELLPPEKYM
RVASPSRKVQFNQNTNLRVIQKNQEETTSISKITVNPDSEELFSDNENNFVFQVANERNN
LALGNTKELHETDLTCVNEPIFKNSTMVLYGDTGDKQATQVSIKKDLVYVLAEENKNSVK
QHIKMTLGQDLKSDISLNIDKIPEKNNDYMNKWAGLLGPISNHSFGGSFRTASNKEIKLS
EHNIKKSKMFFKDIEEQYPTSLACVEIVNTLALDNQKKLSKPQSINTVSAHLQSSVVVSD
CKNSHITPQMLFSKQDFNSNHNLTPSQKAEITELSTILEESGSQFEFTQFRKPSYILQKS
TFEVPENQMTILKTTSEECRDADLHVIMNAPSIGQVDSSKQFEGTVEIKRKFAGLLKNDC
NKSASGYLTDENEVGFRGFYSAHGTKLNVSTEALQKAVKLFSDIENISEETSAEVHPISL
SSSKCHDSVVSMFKIENHNDKTVSEKNNKCQLILQNNIEMTTGTFVEEITENYKRNTENE
DNKYTAASRNSHNLEFDGSDSSKNDTVCIHKDETDLLFTDQHNICLKLSGQFMKEGNTQI
KEDLSDLTFLEVAKAQEACHGNTSNKEQLTATKTEQNIKDFETSDTFFQTASGKNISVAK
ESFNKIVNFFDQKPEELHNFSLNSELHSDIRKNKMDILSYEETDIVKHKILKESVPVGTG
NQLVTFQGQPERDEKIKEPTLLGFHTASGKKVKIAKESLDKVKNLFDEKEQGTSEITSFS
HQWAKTLKYREACKDLELACETIEITAAPKCKEMQNSLNNDKNLVSIETVVPPKLLSDNL
CRQTENLKTSKSIFLKVKVHENVEKETAKSPATCYTNQSPYSVIENSALAFYTSCSRKTS
VSQTSLLEAKKWLREGIFDGQPERINTADYVGNYLYENNSNSTIAENDKNHLSEKQDTYL
SNSSMSNSYSYHSDEVYNDSGYLSKNKLDSGIEPVLKNVEDQKNTSFSKVISNVKDANAY
PQTVNEDICVEELVTSSSPCKNKNAAIKLSISNSNNFEVGPPAFRIASGKIVCVSHETIK
KVKDIFTDSFSKVIKENNENKSKICQTKIMAGCYEALDDSEDILHNSLDNDECSTHSHKV
FADIQSEEILQHNQNMSGLEKVSKISPCDVSLETSDICKCSIGKLHKSVSSANTCGIFST
ASGKSVQVSDASLQNARQVFSEIEDSTKQVFSKVLFKSNEHSDQLTREENTAIRTPEHLI
SQKGFSYNVVNSSAFSGFSTASGKQVSILESSLHKVKGVLEEFDLIRTEHSLHYSPTSRQ
NVSKILPRVDKRNPEHCVNSEMEKTCSKEFKLSNNLNVEGGSSENNHSIKVSPYLSQFQQ
DKQQLVLGTKVSLVENIHVLGKEQASPKNVKMEIGKTETFSDVPVKTNIEVCSTYSKDSE
NYFETEAVEIAKAFMEDDELTDSKLPSHATHSLFTCPENEEMVLSNSRIGKRRGEPLILV
GEPSIKRNLLNEFDRIIENQEKSLKASKSTPDGTIKDRRLFMHHVSLEPITCVPFRTTKE
RQEIQNPNFTAPGQEFLSKSHLYEHLTLEKSSSNLAVSGHPFYQVSATRNEKMRHLITTG
RPTKVFVPPFKTKSHFHRVEQCVRNINLEENRQKQNIDGHGSDDSKNKINDNEIHQFNKN
NSNQAAAVTFTKCEEEPLDLITSLQNARDIQDMRIKKKQRQRVFPQPGSLYLAKTSTLPR
ISLKAAVGGQVPSACSHKQLYTYGVSKHCIKINSKNAESFQFHTEDYFGKESLWTGKGIQ
LADGGWLIPSNDGKAGKEEFYRALCDTPGVDPKLISRIWVYNHYRWIIWKLAAMECAFPK
EFANRCLSPERVLLQLKYRYDTEIDRSRRSAIKKIMERDDTAAKTLVLCVSDIISLSANI
SETSSNKTSSADTQKVAIIELTDGWYAVKAQLDPPLLAVLKNGRLTVGQKIILHGAELVG
SPDACTPLEAPESLMLKISANSTRPARWYTKLGFFPDPRPFPLPLSSLFSDGGNVGCVDV
IIQRAYPIQWMEKTSSGLYIFRNEREEEKEAAKYVEAQQKRLEALFTKIQEEFEEHEENT
TKPYLPSRALTRQQVRALQDGAELYEAVKNAADPAYLEGYFSEEQLRALNNHRQMLNDKK
QAQIQLEIRKAMESAEQKEQGLSRDVTTVWKLRIVSYSKKEKDSVILSIWRPSSDLYSLL
TEGKRYRIYHLATSKSKSKSERANIQLAATKKTQYQQLPVSDEILFQIYQPREPLHFSKF
LDPDFQPSCSEVDLIGFVVSVVKKTGLAPFVYLSDECYNLLAIKFWIDLNEDIIKPHMLI
AASNLQWRPESKSGLLTLFAGDFSVFSASPKEGHFQETFNKMKNTVENIDILCNEAENKL
MHILHANDPKWSTPTKDCTSGPYTAQIIPGTGNKLLMSSPNCEIYYQSPLSLCMAKRKSV
STPVSAQMTSKSCKGEKEIDDQKNCKKRRALDFLSRLPLPPPVSPICTFVSPAAQKAFQP
PRSCGTKYETPIKKKELNSPQMTPFKKFNEISLLESNSIADEELALINTQALLSGSTGEK
QFISVSESTRTAPTSSEDYLRLKRRCTTSLIKEQESSQASTEECEKNKQDTITTKKYI
Equivalence File Format
The equivalence file has very simple format. Every line is considered by the algorithm and all characters per line are considered equivalent. Each character can appear in multiple lines but once per line. Each line must be unique. The input must be in ASCII format (.txt files with rich text option turned off). A sample equivalence file named “SampleEquivalencesTJU.txt” that is included in the downloadable files is the following.
View sample equivalence SampleEquivalencesTJU.txt fileYW
STY
ILV
Output File Format
The output is automatically formatted by the algorithm. It consists of a header that spans 5 lines and 3-4 columns. The results are presented in those 3-4 columns and are as follows:
- Number of pattern occurrences in the entire file
- Number of sequences the pattern appears in
- The pattern
- The list of the position the patterns appears. The first number denotes the sequence and the second the position (This column appears only if -p is set)
So, for example. If we run the algorithm with the input given above and the following parameters
./teiresias -l4 -w6 -k4 -iSampleInputTJU.txt -oMyOutputFile.txt -p
we get the following output.
View the contents of MyOutputFile.txt file##########################################################
# #
# FINAL RESULTS #
# #
##########################################################
5 1 F..ASGK 1 1427 1 1523 1 1843 1 1977 1 2057
5 1 AS.K.V 1 1526 1 1846 1 1980 1 2060 1 2184
5 1 F.TAS.K 1 1008 1 1427 1 1523 1 1977 1 2057
5 2 A.GK.V 0 1707 1 1526 1 1846 1 1980 1 2060
4 1 F..ASGK.V 1 1523 1 1843 1 1977 1 2057
4 1 F.TASGK 1 1427 1 1523 1 1977 1 2057
4 2 E.GSS 0 1045 0 1052 0 1059 1 2138
4 2 LFSD 0 1569 1 881 1 1239 1 2807
4 2 EL.SD 0 180 0 1338 1 880 1 1464
4 2 GSD.S 0 182 0 393 1 1337 1 2440
4 2 SL..AK 0 216 1 1684 1 2508 1 3230
4 2 IKE..S 0 30 0 1158 1 1379 1 3390
Execution examples and what to use when
Every time you run the algorithm you have to specify values for -l,-w,-k and -i. All the other parameters can be used independently and in any combination. The order of the parameters doesn’t matter. When setting your parameters try to think how you would like your results to be and select respectively. Below we provide a list of possible desired results and the recommended parameters. Please keep in mind that even more combinations are possible! When unsure about what to use or how you expect from the data to behave, it might be beneficial to set a parameter very high or very low and then gradually tune it until you get the results you are looking for.
- How to use -i and -o.
Both parameters should be always followed by a file name. The input file parameter, -i, is necessary and the output file parameter -o is optional. In case -o is not given then the name of the output file will be “output.txt”. If you intend to run Teiresias multiple times at a time, you should provide a different name each time for the output files, the output.txt will be created once and overwritten every time you run the program. An usage example is the following:
./teiresias -l2 -w6 -k2 -p -iSampleInputTJU.txt -oOutputBRCA1_BRCA2_HUMAN.txt
- How to use -l and -w.
The parameter -l controls the minimum number of literals in any pattern which is also the minimum length. The parameter -w controls the number of literals that are indifferent, i.e. the number of wildcards, among every -l consecutive (but not necessarily contiguous) literals. It entails that -w must be larger or equal to -l and that -l/-w shows the literal density.
- If you are interested in seeing only exact patterns, in other words -l/-w = 1, you should set -l=-w= the minimum length you wish your smaller pattern to have. For example
./teiresias -l6 -w6 -k4 -iSampleInputTJU.txt
returns patterns that have length at least 6 and all positions have literals.View output file
##########################################################
# #
# FINAL RESULTS #
# #
##########################################################
2 1 NEVGSS
2 1 FSTASGK
2 1 SFSKVI
2 1 SLQNAR
- If you are interested in patterns with wildcards, then you should set -w larger than -l. If -w = -l + 2 for example then each produced pattern will have only two wild cards among every -l consecutive (but not necessarily contiguous) literals. For example
./teiresias -l4 -w6 -k4 -iSampleInputTJU.txt
returns all patterns with length at least 4 that can have (-w)-(-l) = 2 wildcards (‘.’) among every 4 consecutive (but not necessarily contiguous) literals and appear at least 4 times in the input file.View output file
##########################################################
# #
# FINAL RESULTS #
# #
##########################################################
5 1 F..ASGK
5 1 AS.K.V
5 1 F.TAS.K
5 2 A.GK.V
4 1 F..ASGK.V
4 1 F.TASGK
4 2 E.GSS
4 2 LFSD
4 2 EL.SD
4 2 GSD.S
4 2 SL..AK
4 2 IKE..S
- If you are interested in seeing only exact patterns, in other words -l/-w = 1, you should set -l=-w= the minimum length you wish your smaller pattern to have. For example
- How to use -p.
This parameter is quite straightforward. If set then the positions that each pattern appears in the file will be added in the output. For example
./teiresias -l6 -w6 -k4 -iSampleInputTJU.txt -p
returns the positions of all appearances after each pattern.View output file
##########################################################
# #
# FINAL RESULTS #
# #
##########################################################
2 1 NEVGSS 0 1044 0 1051
2 1 FSTASGK 1 1977 1 2057
2 1 SFSKVI 1 1785 1 1868
2 1 SLQNAR 1 1991 1 2482
- How to use -s.
The algorithm is executed in two stages. The scanning phase and the convolution. During the scanning phase all patterns that satify the minimum requirements are found and during the convolution the “elementary” patterns are combined and maximized. When -s is used the algorithm stops after the scanning phase. Although the output patterns are not maximized when this parameter is used, the results are returned much faster and include results that are length specific, as seen below.
If you want to find only the patterns with a specific number of literals you should set -l to that length and use -s. For example:
./teiresias -l5 -w6 -k2 -iSampleInputTJU.txt -s
returns all patterns with exactly 5 literals. If you wanted length 5 also you should set -w=-l=5.View output file
##########################################################
# #
# FINAL RESULTS #
# #
##########################################################
2 1 SKSKS
2 1 LQNAR
2 1 SLQNA
2 1 STASG
2 1 FSTAS
2 1 FSKVI
2 1 SFSKV
2 1 GQPER
4 1 TASGK
2 1 LPRIS
2 1 KEQGL
2 1 RKPSY
2 1 LGPIS
2 2 SLFSD
2 2 LEESG
2 1 EVGSS
2 1 NEVGS
2 1 VQTVN
2 2 ISLLE
2 1 SSSIS
2 2 GSDDS
2 2 GSDSS
2 1 MEKT.S
2 1 SLQN.R
2 1 STAS.K
2 1 FSTA.G
2 1 VSKI.P
2 1 SFSK.I
2 1 KEQG.S
4 1 ASGK.V
2 1 GTGN.L
2 1 LFSD.E
2 1 ETEA.E
2 1 ECEK.K
2 1 EANA.E
2 1 PTLS.T
2 2 LEEN.Q
2 1 EENL.S
2 2 IKED.S
2 2 QTVN.D
2 1 VGSS.N
2 1 NEVG.S
2 1 LGSD.S
2 1 SLQ.AR
2 1 STA.GK
2 1 FST.SG
2 1 SFS.VI
2 1 SDE.YN
2 1 EKE.AK
2 1 NSL.ND
2 1 GSD.SK
2 1 SLN.DK
2 1 LAL.NT
2 1 FSD.EN
2 1 SDT.FQ
2 1 KKV.DI
2 1 KDF.TS
2 2 EEQ.LE
2 2 KNT.EN
2 1 GSS.NE
2 1 NEV.SS
2 1 INE.GS
2 2 ENV.KE
2 2 SPK.TF
2 1 SGL.PE
2 2 EKI.LL
2 2 KKE.NS
2 1 SL.NAR
2 1 ST.SGK
2 1 FS.ASG
2 1 SF.KVI
2 1 LK.SKS
2 1 SL.KVK
2 1 GF.TAS
2 1 KI.KES
2 1 QL.ATK
2 1 SD.SKN
2 1 SV.VSD
2 1 KE.KLS
2 1 SQ.SLL
2 2 TS.LPR
2 2 EQ.TSE
2 2 DK.LVS
3 1 NE.GSS
2 2 LS.ERV
2 2 SK.HDS
2 1 SD.SED
2 2 KE.NSP
2 1 S.QNAR
2 1 S.ASGK
2 1 S.SKVI
2 1 L.TSKS
2 1 F.GQPE
4 1 F.TASG
2 1 K.AGLL
2 1 K.NNYE
2 2 S.VKEL
2 2 D.RNPE
2 1 S.LEDL
2 1 V.NIPS
2 2 E.LSSE
2 2 L.SDNL
2 1 N.VGSS
2 2 E.SSSN
2 2 E.FEEH
2 2 S.ERVL
2 1 V.SKSV
2 2 L.NQKK
2 2 E.IKEP
- How to use -k, -q and -v.
The parameter -k controls the minimum number of appearances a pattern can have. The parameter -q controls the maximum. The parameter -v controls how the appearances are counted. If omitted then they are counted per general appearance but if included each pattern is counted once per sequence. There are many interesting combinations among those variables.
- If you are looking for patterns that appear at least -k times or/and no more than -q, set -k and -q accordingly. For example:
./teiresias -l3 -w3 -k5 -q5 -iSampleInputTJU.txt
returns all exact patterns that span at least 3 positions and appear exactly 5 times.View output file
##########################################################
# #
# FINAL RESULTS #
# #
##########################################################
5 2 TVN
5 2 SLK
5 2 LPR
5 2 VEE
5 2 NVE
5 2 GLE
5 2 KTK
5 2 SEN
5 2 SNN
5 2 GKE
5 2 KEL
5 2 LSN
5 2 LES
5 2 ENI
5 2 GLL
5 2 GSD
5 2 QSP
5 2 SCS
5 2 TKC
5 2 SET
5 2 ENK
5 2 ESS
5 2 KKK
5 2 SDE
5 2 EKN
5 2 PTS
5 2 VST
5 2 TSL
5 2 STR
5 1 KVK
5 1 ASGK
5 1 RAL
5 1 KLS
5 2 ESG
5 2 SST
5 1 SFS
5 2 SLP
5 1 DSE
5 2 EES
5 2 SLN
5 2 EIK
5 2 SQK
5 2 PIS
5 2 SRN
5 2 SEL
5 2 ETV
5 2 TEN
5 2 DSS
5 2 FSD
5 2 SIS
5 2 KCS
5 2 SVV
5 2 SIK
5 2 LKN
5 2 SLF
5 2 SSA
5 2 EEQ
5 2 AKK
- If you want to find patterns that exist in a specific number of sequences (or all) then set -v and -k accordingly. For example
./teiresias -l4 -w4 -k2 -v -iSampleInputTJU.txt
returns all patterns with length 4 or more and no wildcards that appear in both the sequences of the sample input file.View output file
##########################################################
# #
# FINAL RESULTS #
# #
##########################################################
4 2 LFSD
3 2 NEVG
3 2 QTVN
3 2 SKNK
3 2 SLQN
3 2 SDSS
3 2 LEEN
3 2 SLFS
3 2 SLLE
3 2 IKKK
3 2 SSED
2 2 LSNL
2 2 PEKY
2 2 AEND
2 2 LSKN
2 2 SDNL
2 2 HLSE
2 2 NSLN
2 2 SNSY
2 2 TSVS
2 2 RNPE
2 2 SDTF
2 2 KSVQ
2 2 VFSK
2 2 LESS
2 2 TEQN
2 2 ITPQ
2 2 CNKS
2 2 LEESG
2 2 GSKE
2 2 SLPR
2 2 FGKT
2 2 SSKQ
2 2 TLKY
2 2 DTYL
2 2 SEET
2 2 IKED
2 2 SPAA
2 2 GSSE
2 2 VEEL
2 2 ASTE
2 2 FTKC
2 2 KENN
2 2 VGQK
2 2 FKTK
2 2 FEEH
2 2 ERVL
2 2 PEHL
2 2 SVSE
2 2 GSDDS
2 2 EQKE
2 2 EEKE
2 2 DEEL
2 2 ISLLE
2 2 KSER
2 2 EISL
2 2 ESTR
2 2 LIKE
2 2 NQKK
2 2 KCSI
2 2 SPYL
2 2 KVSK
2 2 TENL
2 2 GSDSS
2 2 APES
2 2 SLFSD
2 2 SISN
2 2 SEEI
2 2 SVVK
2 2 SETS
2 2 SSSN
2 2 NVEK
2 2 IKEP
2 2 KTSV
2 2 NKSK
2 2 ENNS
2 2 SPER
2 2 ELSS
2 2 VKEL
2 2 VKTK
2 2 EETT
2 2 EESE
2 2 DLLD
2 2 SQKS
2 2 SLKK
2 2 VHPI
2 2 KADL
2 2 NQEE
2 2 DIKE
2 2 SVAL
2 2 GLEI
2 2 VNKR
2 2 LSSE
2 2 EAAS
2 2 LGRN
2 2 ETSY
- If you are looking for patterns that appear at least -k times or/and no more than -q, set -k and -q accordingly. For example:
- How to use -b and -n.
The parameter -n controls the number of bracketed literals, it should always be used with -b. These parameters are used when some amount of freedom of replacement is given to the literals but the replacements can only occur within specific groups. Of course they can also be combined with wildcards. The result will be patterns with some specific literals, some literals that can only be chosen among the bracketed ones and some wildcards i.e. literals that can be anything. Keep in mind that unlike -w that signifies the maximum number of ‘.’ among every -l consecutive (but not necessarily contiguous) literals, -n signifies the number of brackets per pattern. If -n is not set but a file is provided using -b, then -n is set to “infinity”. The brackets are trimmed so that only literals that actually appear in the input in those sequences remain.
- If you don’t want to use wildcards but would still like some positions to be non specific, you should use -b. For example
./teiresias -l4 -w4 -k5 -iSampleInputTJU.txt -bSampleEquivalencesTJU.txt
returnsView output file
##########################################################
# #
# FINAL RESULTS #
# #
##########################################################
11 2 [STY][STY][ILV][ILV]
11 2 [ST][STY][ILV][SY]
10 2 [ST][STY][ILV]S
9 2 [ILV][STY][ST][ILV]
9 2 [ILV][ILV][ST][STY]
8 2 [IV][SY][ILV][ILV]
8 2 [ST][ST][STY][ILV]
8 2 [ILV][STY][ILV][ST]
8 2 [ST]S[ILV][SY]
7 2 [ST]S[ILV]S
7 2 [ILV][ILV][STY][ILV]
7 2 [ILV]K[ILV][ST]
7 2 [ILV][STY][STY][STY]
7 2 [ST][ILV][ST][ST]
7 2 [ST][ILV][ILV]E
7 2 [IV]S[ILV][ILV]
6 2 [ILV][ILV][SY]D
6 2 S[STY][ILV][ILV]
6 1 [ILV][STY][ILV]S
6 2 [IL]F[ST]D
6 2 D[ILV][ILV][STY]
6 2 [ILV][STY]L[ST]
6 2 T[SY][ILV][SY]
6 2 [ILV][STY]E[ST]
6 2 EE[ILV][ILV]
6 2 [STY][ILV][ILV][STY]
6 2 [ST]S[STY][ILV]
6 2 [ILV]S[ST][IL]
6 2 [ILV]K[ILV]Sv
6 2 [ILV][STY][ST]L
6 2 K[ST][ST][ST]
6 2 [ST][ILV][SY][ILV]
6 2 E[ILV][STY][STY]
6 2 S[ILV][ILV]E
6 2 [ST][LV][ILV][ILV]
6 2 [STY][STY][ILV]L
6 2 [ST][STY]EE
6 2 [ILV]D[ILV][IL]
6 2 [STY][STY]L[IL]
6 2 [STY]S[LV][ILV]
5 2 [ILV][ILV]KE
5 2 V[SY][ILV][ILV]
5 2 [STY][ILV][ST]N
5 2 TS[ILV][SY]
5 2 [ST][ST]V[SY]
5 2 [SY][ILV]E[ST]
5 2 [ST][STY][SY]K
5 2 [SY]G[ILV][STY]
5 2 [LV]E[STY]S
5 2 E[ST]D[ILV]
5 2 LF[ST]D
5 2 [ILV][ST]S[ST]
5 2 [ILV][ILV]T[ST]
5 2 SN[ST][STY]
5 2 [LV][ILV][ILV]Q
5 2 [ST][IL]KK
5 1 [IL]S[ILV][ST]
5 2 [ST][STY][STY][SY]
5 2 [ILV][ILV]SD
5 2 [LV][ST][STY]E
5 1 [ST]ASG
5 1 S[ST][IV]S
5 1 ASGK
5 2 [ST][ST]S[IL]
5 2 [ST]D[ILV][STY]
5 2 [LV][ILV][LV][ST]
5 2 [ST][IL]LE
5 2 [ILV][ST][SY]S
5 2 S[ILV][SY][ILV]
5 2 [IL][STY][STY]Q
5 2 K[ST]S[ST]
5 2 [ST][LV]S[ST]
5 2 [LV][STY][IL]E
5 2 [ILV][ST]T[ILV]
5 2 DE[ILV][STY]
5 2 [ILV][ST]N[ST]
5 2 E[ST][ST][SY]
5 2 [SY][IL]QN
5 2 H[ILV][STY]E
5 2 S[LV]F[ST]
5 2 [ST]G[IL]E
5 2 [ILV][ILV]S[ILV]
5 2 [STY]P[STY][ST]
5 2 [ILV][ST]N[LV]
5 2 T[SY][ILV]S
- The number of brackets per pattern can be restricted by using -n. For example
./teiresias -l4 -w4 -k5 -n2 -iSampleInputTJU.txt -bSampleEquivalencesTJU.txt
returnsView output file
##########################################################
# #
# FINAL RESULTS #
# #
##########################################################
7 2 [ST]S[ILV]S
6 2 [ST][STY]EE
6 2 S[ILV][ILV]E
6 2 [ILV]K[ILV]S
6 2 EE[ILV][ILV]
6 2 [IL]F[ST]D
5 2 [ILV][ILV]KE
5 2 [ST][IL]LE
5 2 K[ST]S[ST]
5 2 DE[ILV][STY]
5 2 [SY][IL]QN
5 2 H[ILV][STY]E
5 2 S[LV]F[ST]
5 2 [ST]G[IL]E
5 2 T[SY][ILV]S
5 2 TS[ILV][SY]
5 2 [LV]E[STY]S
5 2 E[ST]D[ILV]
5 2 LF[ST]D
5 2 SN[ST][STY]
5 2 [ST][IL]KK
5 2 [ILV][ILV]SD
5 1 [ST]ASG
5 1 S[ST][IV]S
5 1 ASGK
- If you don’t want to use wildcards but would still like some positions to be non specific, you should use -b. For example
How to download and run
Instructions for Unix/MACOS- Download the compressed file from here.
- Right click on the file and un-compress it or open it. Move the folder created on your desktop or the files in a folder on your desktop.
- Open a terminal window. To do that, go to the launchpad and in the search box write “terminal”, then click on the terminal.
- On the terminal, type “cd Desktop/name-of-folder/” where name-of-folder is the name of the folder on the desktop containing the files. Press enter.
- Type “make”. Press enter.
- Type “./teiresias -l4 -w6 -k4 -iSampleInputTJU.txt -p” or any other parameters you want and press enter.
- Open the output file created in the same folder on the desktop to see the results!
Instructions for Windows/DOS
Coming soon.
Changelog
v0.9.1 UPDATED 3/19/2014- ReadMe file added
- The input file can contain empty lines
- The input file’s headers can contain anything (must still start with >)
- The input file’s sequences can have end-of-line characters among their characters
Web App
Input options and format
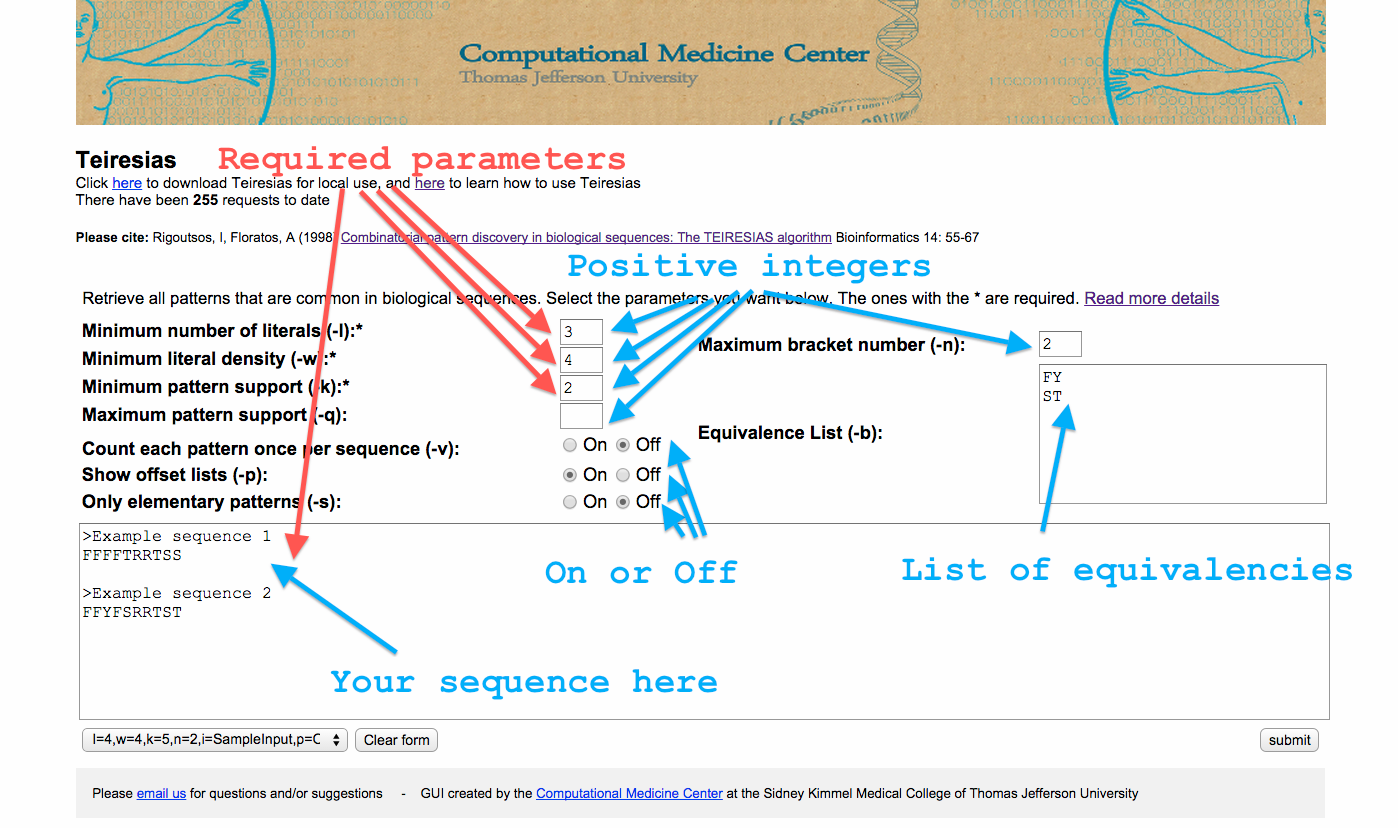
The Teiresias web-app requires the same parameters as the source code. You can read more information about what each parameter is here. The required parameters are l,w, and k. In short, when a pattern is returned from Teiresias it will have the form, abcdefgh. Some of those literals (here the letters a-h) will be specified (here a letter between a-h) and others will be wildcards (a range of letters). The parameter l defines the number of literals that have to be specified. The first and last literal are always specified, so l must be at least 2. The result of w-l defines how many literals can be wildcards among every w input characters. Hence, w needs to be at least 2. And finally k is the number of times a pattern has to at least appear in order to be reported.
In the web app you’ll need to add a positive integer in the boxes beside l,w, k and if you want q and n. And optionally select a value for v, p, and s by clicking on “on” or “off”. Also you will need to add your input and optionally an equivalence list. You can find the required formats for the input and equivalence boxes can be found here.
You can also select “Click here for sample parameters” to see an example of the parameters and the results of the app. See the image below for more details.
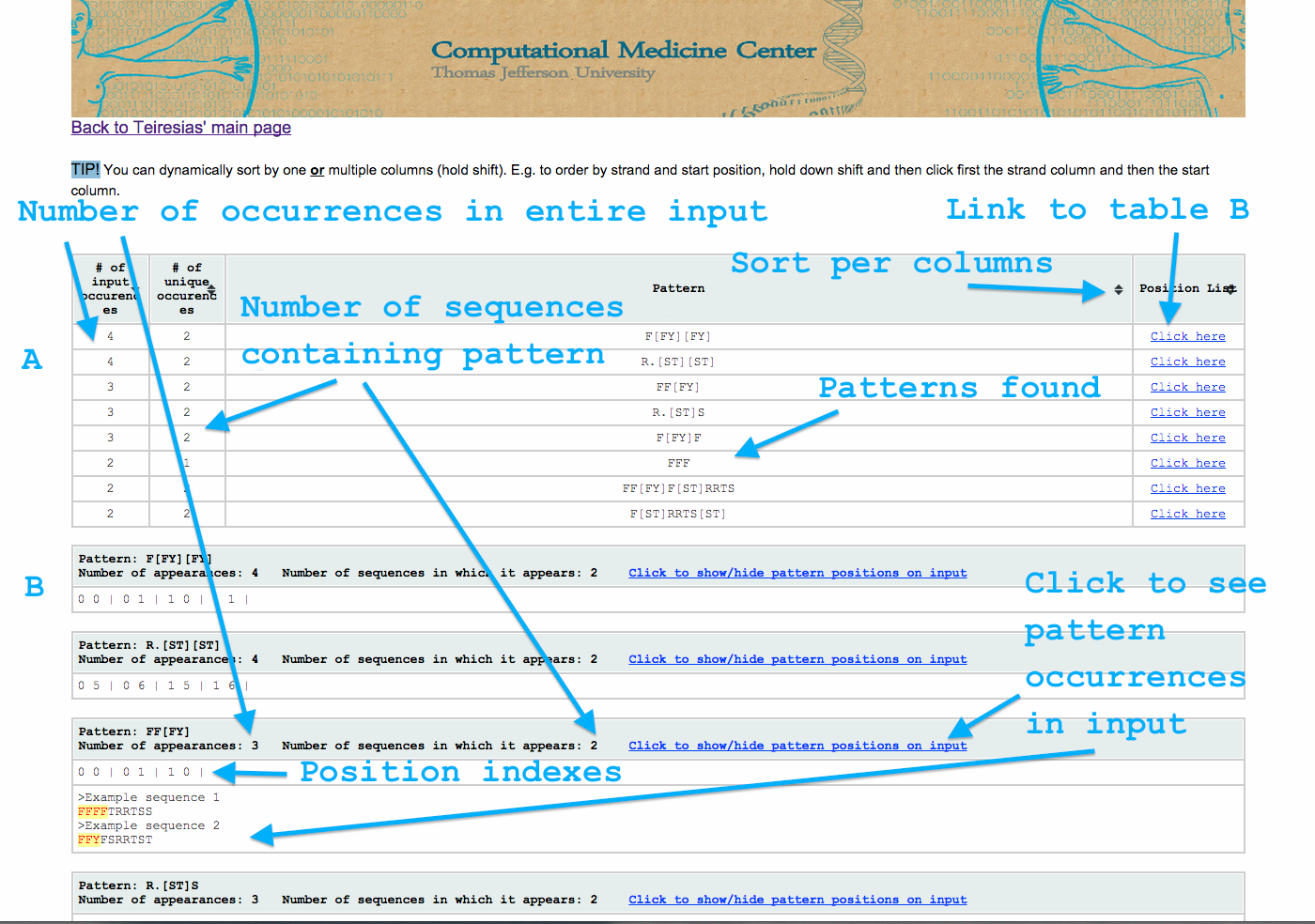
Output options and layout
The output is divided into 2 sections (see image below). The top of the page (table A) contains a summary of the results. The bottom contains a collection of tables that contain the detailed results per pattern.
Table A has 4 columns.
- The number of occurrences of the pattern in the entire input (see also parameters k and q)
- The number of the sequences containing the pattern (see also parameter v)
- Pattern (see parameters l,w,s,n,i and b)
- Link to individual table (table B)
Table B is a collection of tables. Each has 3 lines.
- The first line contains:
- The pattern (see parameters l,w,s,n,i and b)
- The number of total appearances (see also parameters k and q)
- The number of sequences the pattern appears on (see also parameter v)
- link that opens the third line.
- The second line contains the list of coordinates in which the patterns appears within the input. Each coordinate consists of 2 numbers and is followed by a bar “|”. The first number is the sequence in which the pattern is and the second is the character at which it starts (see also parameter p).
- The third line is optional and depicts the pattern’s appearances at the input.
You can also sort the contents of table A by one or multiple columns.
References
- Rigoutsos, I, Floratos, A (1998) Combinatorial pattern discovery in biological sequences: The TEIRESIAS algorithm. Bioinformatics 14: 55-67
You are visitor number 3417.