We describe the profile of tRNA-derived fragments (tRFs) in more than 700 individuals and two different tissues: Lymphoblastoid cell lines (LCL) and breast cancer (BRCA) and normal breast tissue. Our in-depth and detailed computational analysis revealed a novel distinct category of tRFs, the i-tRFs that are wholly internal to the mature tRNA molecule. We provide strong evidence that the i-tRFs as well as the known categories of 5’-tRFs and 3’-tRFs are constitutive in nature. We additionally identify specific dependencies of the tRF profile: it is different for the two genders and also different in different human races. tRFs are not only tissue-specific, as the LCL and the BRCA datasets expressed different fragments, but also tissue-state specific, as the tRF expression significantly changed from normal to tumor tissue. Importantly, different disease subtypes exhibited differences in their tRf profiles. These molecular characteristics have important implications in personalized medicine, as they lay the groundwork for subsequent studies on studying human racial variation deciphering health disparities.
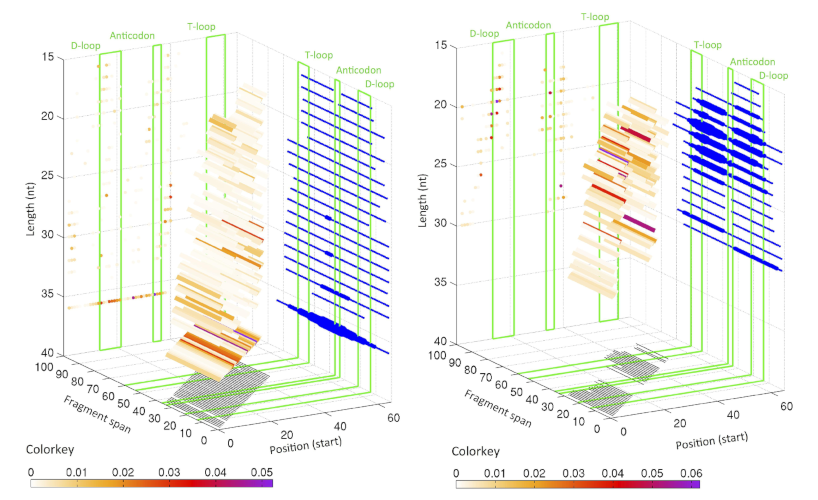 Distribution of starting position and lengths for i-tRFs for LCL and BRCA datasets
Distribution of starting position and lengths for i-tRFs for LCL and BRCA datasets
Read More
You can also see the datasets used in the study.
References
- Telonis, AG, Loher, P, Honda, S, Jing, Y, Palazzo, J, Kirino, Y, Rigoutsos, I. Dissecting tRNA-derived fragment complexities using personalized transcriptomes reveals novel fragment classes and unexpected dependencies. Oncotarget. 2015;6 (28):24797-822. doi: 10.18632/oncotarget.4695. PubMed PMID:26325506 PubMed Central PMC4694795.
