After transcription, newly-synthesized primary RNAs undergo various maturation steps prior to becoming functional molecules. Post-transcriptional non-template additions of nucleotides to 3′-ends of RNAs represent a major maturation step that regulates the stability and function of RNA molecules. Yet, RNAs containing additional 3′-terminal sequences are expected to be infrequently recognized and characterized in RNA-seq data, because the presence of non-genomic sequences reduces their chances of being mapped on the genome. Considering the relative obscurity of these RNAs, cells may express many yet-unidentified RNA molecules with post-transcriptionally-added non-genomic sequences on their 3′-ends.
Transfer RNAs (tRNAs) are universally expressed non-coding RNAs (ncRNAs) that play central roles in translation machinery. Their 3′-CCA end is not genetically encoded and is post-transcriptionally added by tRNA nucleotidyltransferase (CCA-adding enzyme). In addition to tRNAs, other ncRNAs can be substrates of CCA-adding enzyme, but endogenous non-tRNA substrates of CCA-adding enzyme have not yet been widely explored.
We previously developed a Y-shaped Adapter-ligated MAture TRNA sequencing (YAMAT-seq) that efficiently and conveniently allows high-throughput sequencing of tRNAs (Shigematsu et al., NAR 2017). YAMAT-seq captures RNA species that form secondary structures with 4-nucleotide protruding 3′-ends of the sequence 5′-NCCA-3′, which is the hallmark structure of RNAs that are generated by CCA-adding enzyme. We therefore used YAMAT-seq to identify non-tRNA substrates of CCA-adding enzyme.
By executing YAMAT-seq for human breast cancer cells and mining the sequence data, we identified novel candidate substrates of CCA-adding enzyme. These included fourteen “CCA-RNAs” that only contain CCA as non-genomic sequences, and eleven “NCCA-RNAs” that contain CCA and other nucleotides as non-genomic sequences. All newly-identified (N)CCA-RNAs were derived from the mitochondrial genome and were localized in mitochondria. Knockdown of CCA-adding enzyme severely reduced the expression levels of (N)CCA-RNAs, suggesting that the CCA-adding enzyme-catalyzed CCA additions stabilize the expression of (N)CCA-RNAs. Furthermore, expression levels of (N)CCA-RNAs were severely reduced by various cellular treatments, including UV irradiation, amino acid starvation, inhibition of mitochondrial respiratory complexes, and inhibition of the cell cycle. These results revealed a novel CCA-mediated regulatory pathway for the expression of mitochondrial ncRNAs.
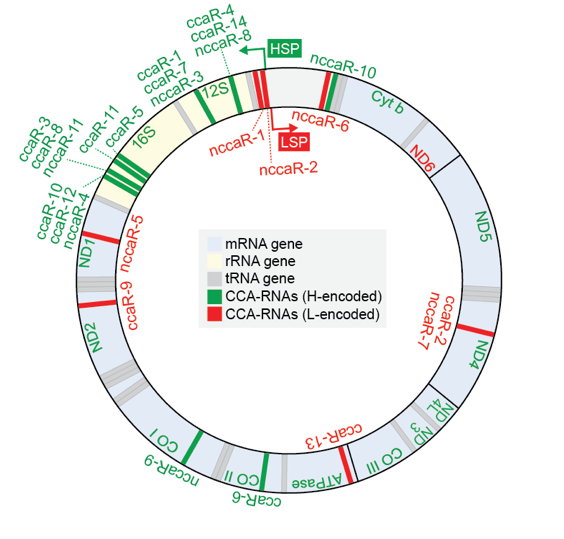 YAMAT-seq data reveals a new class of mitochondrial RNAs, CCA-RNAs
YAMAT-seq data reveals a new class of mitochondrial RNAs, CCA-RNAs
References
- Pawar, K, Shigematsu, M, Loher, P, Honda, S, Rigoutsos, I, Kirino, Y. Exploration of CCA-added RNAs revealed the expression of mitochondrial non-coding RNAs regulated by CCA-adding enzyme. RNA Biol. 2019;16 (12):1817-1825. doi: 10.1080/15476286.2019.1664885. PubMed PMID:31512554 PubMed Central PMC6844566.
