Cells express immensely diverse species of RNA molecules that play essential roles in numerous biological processes. With the advent of next-generation sequencing (NGS) technology, efforts to identify the expressed RNA molecules have greatly advanced our understanding of RNA biology. Although NGS-based RNA-sequencing (RNA-seq) has become a ubiquitous tool in biological and medical research, the current standard RNA-seq methods, particularly those targeting short non-coding RNAs (ncRNAs), do not fully capture all of the RNAs expressed but allow for some “escapers” to slip by undetected. RNAs generated by ribonuclease cleavage sometimes harbor a 2′,3′-cyclic phosphate (cP) at their 3′-ends, and those cP-containing RNAs (cP-RNAs) are one such escaper that are not ligated to a 3′-adapter and thus uncaptured by standard RNA-seq.
An increasing number of studies has been suggesting biological significances of the expression of cP-RNAs. Yet, cP-RNAs remained a hidden component in the transcriptome, infrequently recognized and characterized. Given that cP-RNAs are expressed as functional molecules, capturing the entire repertoire of cP-RNAs would broaden the catalog of functional ncRNAs and reveal significant biological events that have been eluding standard RNA-seq. We previously developed “cP-RNA-seq,” which is able to specifically sequence cP-RNAs (Honda et al., PNAS 2015; Honda et al., Nat Protoc 2016).
Here we provide first genome-wide analyses of short cP-RNA transcriptome across multiple mouse tissues. Using cP-RNA-seq that can exclusively sequence cP-RNAs, we identified numerous novel cP-RNA species which are mainly derived from cytoplasmic transfer RNAs (tRNAs), messenger RNAs (mRNAs), and ribosomal RNAs (rRNAs). Determination of the processing sites of substrate RNAs for cP-RNA generation revealed highly-specific RNA cleavage events between cytidine and adenosine in cP-RNA biogenesis. cP-RNAs were not evenly derived from the overall region of substrate RNAs but rather from specific sites, implying that cP-RNAs are not from random degradation but are produced through a regulated biogenesis pathway. The identified cP-RNAs were abundantly accumulated in mouse tissues, and the expression levels of cP-RNAs showed age-dependent reduction. These analyses of cP-RNA transcriptome unravel a novel, abundant class of non-coding RNAs whose expression could have physiological roles.
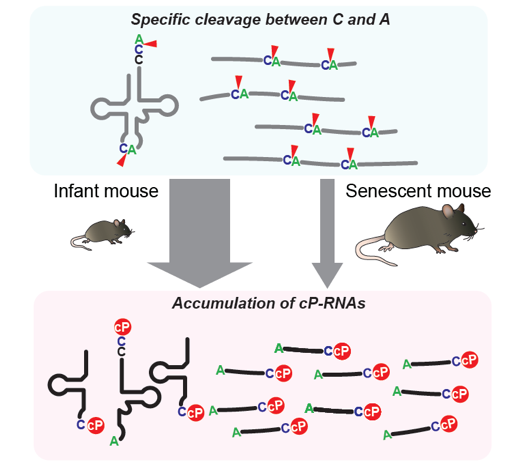 Generation of cP-RNAs and its age-dependent decrease
Generation of cP-RNAs and its age-dependent decrease
Read More
This paper was also covered on EurekAlert!
References
- Shigematsu, M, Morichika, K, Kawamura, T, Honda, S, Kirino, Y. Genome-wide identification of short 2',3'-cyclic phosphate-containing RNAs and their regulation in aging. PLoS Genet. 2019;15 (11):e1008469. doi: 10.1371/journal.pgen.1008469. PubMed PMID:31721758 PubMed Central PMC6853296.
