#OpenScience #OpenCode #OpenData #MachineLearning
Blog Summary
I had the privilege to be a project sponsor and mentor for a sharp group of undergraduate and graduate students taking an AI Computer Vision course at UNCC (the University of North Carolina at Charlotte). These students applied advanced machine learning techniques leveraging ConvNets (convolutional neural networks) from fast.ai, to successfully classify classes of RNA molecules that our center studies. This framework shows the potential for image classifiers to identify de novo molecules and to distinguish those that may be less functional (e.g. pseudo tRNA).
Background
As part of a computer vision course at UNCC taught by Professor Stephen Welch, students were assigned an industry partner who presented them with a problem to work on for their semester long course project. I had the privilege to be the project sponsor and help guide the following students: Rishi Koushal, Glenn McCurdy, Kevin Mitchell, and Vinay Krishna.
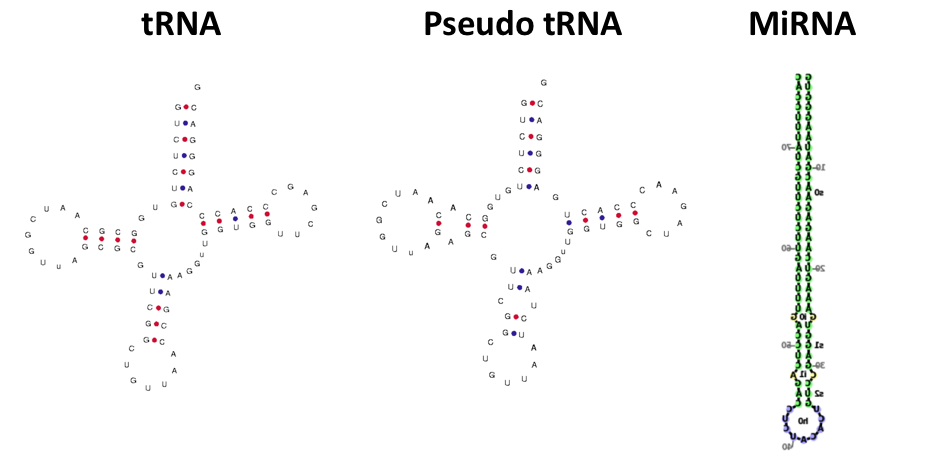
I presented the group with the following questions (1) can an image classifier be used to tell apart different classes of RNA molecules that we study (e.g. miRNAs, tRNAs) and (2) is the image classifier precise enough to distinguish between functional vs non-functional molecules (e.g. pseudo tRNA) molecules. For example, miRNA precursors form a hairpin shape while tRNAs form a cloverleaf. While pseudo tRNA’s often times form a cloverleaf, they can have minute differences in base pairings or shape that prevent it from being functional. Biochemists can often times pick out these differences – so we wanted to see how an image classifier performed.
In summary – the team first generated images from various classes of molecules molecules. They did this by downloading known and annotated molecular sequences from online repositories and leveraged already published RNA folding algorithms to generate images comprising the molecules’ predicted secondary structures. Then they leveraged the fastai library to train ConvNets to perform image classification. Their initial results indicated a promising 90% accuracy.
Perhaps what impressed me the most is how fast this team was able to adapt to a new domain (molecular biology) and the domain-specific complexity present in such data. As advances in artificial intelligence and machine learning continue to make it across domains, the multidisciplinary nature of research will continue to increase. I’m excited about the possibility in leveraging such techniques to help our team discover and better understand new molecules.
Data and Code Availability
The students made their source code and data for this project available on GitHub. Some of the key packages utilized were ViennaRNA (RNA folding), Biopython (FASTA format processing), and Fastai (ConvNets).




