In recent years, the role of microRNAs (miRNAs) as biomarkers in human diseases has been studied due to the function of miRNAs to control and regulate gene expressions. However, research involving the impact of specific miRNAs and their specific influence on diseases is inefficient due to a significant obstacle in the medical industry: the constantly updating PubMed database containing thousands of unsorted and complex abstracts. With the above observation in mind, during the summer of 2020, we, Alexander Liang and Matt Laws, began a six-week internship under Thomas Jefferson University professor, Dr. Nestoras Karathanasis. Our internship focused on the implementation of a text-mining program that employed the R programming language. Our software automatically and efficiently extracts miRNA biomarkers – disease relationships within publicly available abstracts on PubMed.
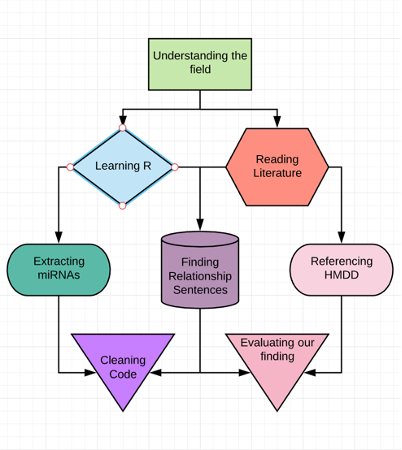 Diagram of the tasks we performed in our internship
Diagram of the tasks we performed in our internship
Per abstract, our program extracts six pieces of important information per abstract:
- diseases: all diseases mentioned in the abstract
- miRNAs: all miRNAs mentioned in the abstract
- relationships: all sentences containing relationships between a disease and miRNA
- PMID: the identifier for each abstract
- organisms: organisms that the miRNAs belong to
- countries: geographic information of research
The end product is a data table with rows containing the succinct summaries of thousands of abstracts, generated at the approximate rate of one summary for an abstract per second. The program compiles the information in a concise, easy to access spreadsheet where each row contains all 6 pieces of extracted information.
Following extraction, we evaluated the accuracy of our tool using the manually curated Human microRNA Disease Database (HMDD) [1] database as our ground truth. We employed three statistics:
- recall, which measures the program’s ability to extract relevant relationships according to HMDD
- precision, which measures the program’s ability to only generate true relationships according to HMDD
- f-score, which averages the previous two statistics
At the end of the internship, the program’s best performance on finding miRNA-disease relationships for a disease was a recall of 0.731, a precision of 0.864, and a f-score of 0.792.
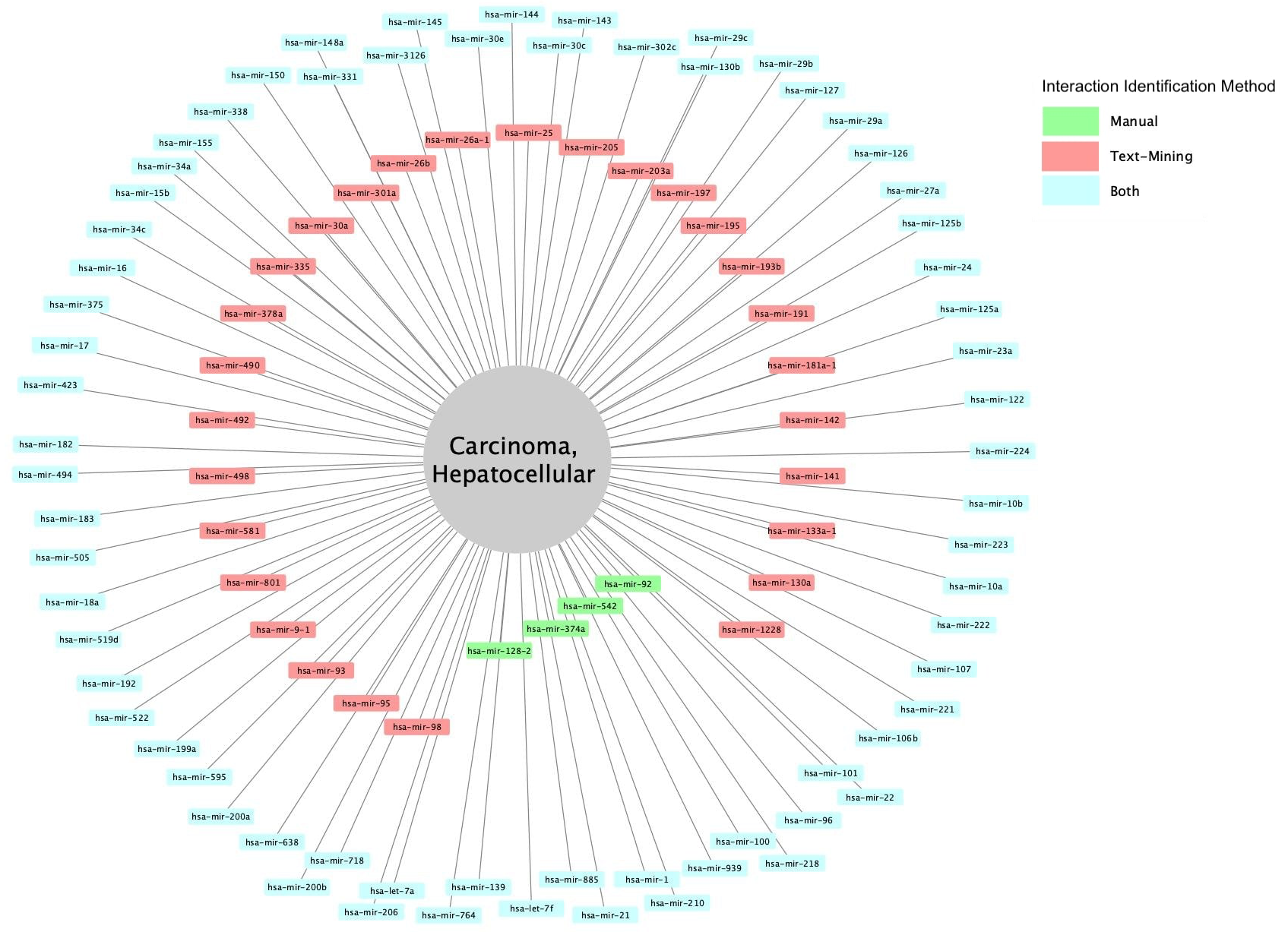 miRNA biomarkers for hepatocellular carcinoma
miRNA biomarkers for hepatocellular carcinoma
Read More
For more information you can download our presentation slides.
References
- Huang, Z, Shi, J, Gao, Y, Cui, C, Zhang, S, Li, J, et al. (2019). HMDD v3.0: A database for experimentally supported human microRNA-disease associations. Nucleic Acids Res. 47, D1013–D1017. doi:10.1093/nar/gky1010. PubMed PMID:30364956.
About Authors

Alexander Liang
Summer Intern
Alex is a senior at the Lawrenceville School in New Jersey. He is interested in computer science, physics, and mathematics. He joined the Jefferson Computational Medicine Center in the summer of 2020 to explore the interdisciplinary study between biology and data science. He is self-taught in multiple languages and is currently doing a study on Physics E&M while also researching further into biophysics.
...Read More
Matt Laws
Summer Intern
I grew up with two Ph.D. chemists as parents and always loved science, picked up coding in the summer of 2019, and have loved it ever since. I have loved to build stuff ever since I was younger, and recently taken up furniture construction.
...Read MoreSupervisor

Nestoras Karathanasis
Teaching Assistant Professor
Dr. Nestoras Karathanasis is a teaching assistant professor at the College of Life Science at Thomas Jefferson University (TJU) in Philadelphia. He received his PhD from the University of Crete, Greece in the field of "miRNA-mRNA interactions related with cancer" in 2013 and in 2015 was appointed Lead Bioinformatician at Miroculus company in San Francisco before joining TJU.
He has an active interest in precision medicine and his work focuses on the statistical integration of different omics data, the development of miRNA mapping tools and apply machine learning algorithms to address biological questions in key areas such as cancer. As a teaching assistant professor, he has developed three graduate-level courses on computational skill acquisition, (i.e. R programming, data visualization, and transcriptomics data analysis). In 2018, he created and led a machine learning team from the Computational Medicine Center of TJU in participating in the DREAM Single Cell Transcriptomics challenge. His team ranked 6th place among 49 participating teams. As of 2019, he is co-recipient of the TJU Data Science Award for his work as a member of the Computational Medicine Center team.
...Read More