It is well established that small noncoding RNAs (sncRNAs) exist in our cells and play an important role in our health. Our previous work with sncRNAs such as microRNAs (miRNAs), miRNA isoforms (isomiRs), and tRNA-derived fragments (tRFs) showed that their production and abundances depend on personal attributes. In this study, we characterize a new class of sncRNAs that are derived from ribosomal RNA (rRNA) and are called rRNA-derived fragments or “rRFs.”
We studied the RNA profiles of immortalized cell lines derived from the B-cells of participants in the 1KG Project. Specifically, we analyzed data obtained from 434 healthy males and females, from five different geographical populations around the world. We focused on small (≤ 33 nucleotides) sncRNAs that mapped to rRNAs. Four such rRNAs are encoded in the nuclear genome whereas two more are encoded in the mitochondrial genome.
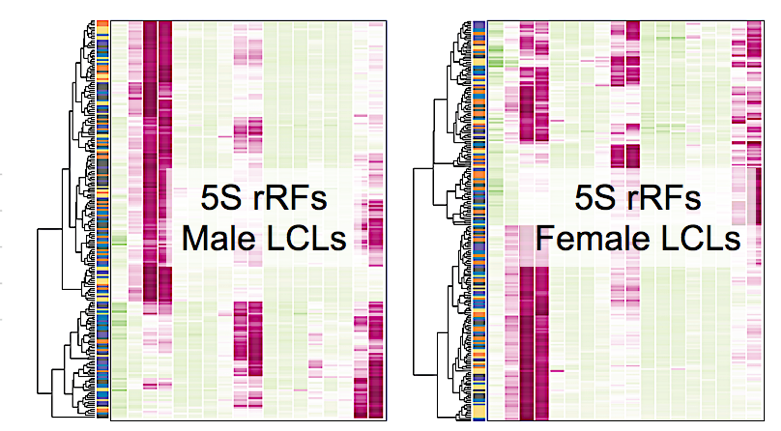
We found that each of the six rRNAs produced rRFs with distinct length and abundance profiles. Also, we demonstrated that while many of these rRFs are produced across all populations and both sexes, the expression levels of a significant number of rRFs are population- and sex-specific. Further analyses of data from additional tissue/cell types showed that the rRF profiles also depend on tissue type, tissue state, and disease type.
 The length distributions of 5S rRFs for five LCL populations from the 1KG Project
The length distributions of 5S rRFs for five LCL populations from the 1KG Project
This study describes a comprehensive analysis of a new class of sncRNAs, the rRFs, which depend on population-, sex-, tissue-, and disease-specific factors. It is still unknown how rRFs are produced, what they do, or how they do it. Nonetheless, the rRF properties that we uncovered mirror our previous findings on the functionally important isomiRs and tRFs. Our detailed characterization of rRFs is an important first step towards prioritizing among the various rRFs and providing a framework in which to study the role that rRFs play in health and disease. You can find more about our analyses here.
rRNA fragment profiles of LCLs from 1000 Genomes Project
We have created the ribosomal RNA fragments (rRFs) expression profiles in the RNA-seq datasets from the lymphoblastoid cell lines (LCLs) of the 1000 Genomes (1KG) Project. You can find the sequences of the 16,279 rRFs that survive the imposed thresholds (Threshold-seq, ≥ 10 RPM, length cutoffs) in Additional file 4 of our publication.
Read More
You can read the Press Release of this paper.
References
- Cherlin, T, Magee, R, Jing, Y, Pliatsika, V, Loher, P, Rigoutsos, I. Ribosomal RNA fragmentation into short RNAs (rRFs) is modulated in a sex- and population of origin-specific manner. BMC Biol 18, 38 (2020). doi: 10.1186/s12915-020-0763-0. PubMed PMID:32279660
